You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000091_05854
You are here: Home > Sequence: MGYG000000091_05854
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Achromobacter xylosoxidans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Achromobacter; Achromobacter xylosoxidans | |||||||||||
| CAZyme ID | MGYG000000091_05854 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 293374; End: 294648 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 110 | 296 | 5.8e-127 | 0.9946524064171123 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00656 | Amb_all | 1.44e-46 | 115 | 295 | 1 | 188 | Amb_all domain. |
| COG3866 | PelB | 1.76e-39 | 75 | 295 | 49 | 275 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam00544 | Pec_lyase_C | 1.17e-22 | 121 | 293 | 27 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CKH39919.1 | 8.97e-312 | 1 | 424 | 1 | 424 |
| SQG72278.1 | 8.97e-312 | 1 | 424 | 1 | 424 |
| CCH08513.1 | 3.26e-307 | 11 | 424 | 1 | 414 |
| AHC47209.1 | 2.37e-294 | 26 | 424 | 1 | 399 |
| AMH07573.1 | 2.19e-287 | 38 | 424 | 1 | 387 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FI2_A | 7.10e-256 | 30 | 424 | 8 | 402 | VexL:A periplasmic depolymerase provides new insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides [Achromobacter denitrificans] |
| 1PXZ_A | 3.77e-26 | 69 | 298 | 27 | 273 | ChainA, Major pollen allergen Jun a 1 [Juniperus ashei],1PXZ_B Chain B, Major pollen allergen Jun a 1 [Juniperus ashei] |
| 3ZSC_A | 1.08e-16 | 76 | 293 | 23 | 226 | Catalyticfunction and substrate recognition of the pectate lyase from Thermotoga maritima [Thermotoga maritima] |
| 3VMV_A | 1.39e-15 | 129 | 295 | 80 | 248 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1PCL_A | 2.55e-14 | 121 | 269 | 73 | 245 | ChainA, PECTATE LYASE E [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FM66 | 5.17e-32 | 69 | 305 | 74 | 323 | Putative pectate lyase 21 OS=Arabidopsis thaliana OX=3702 GN=At5g55720 PE=3 SV=1 |
| P15721 | 1.02e-30 | 69 | 298 | 72 | 317 | Probable pectate lyase P56 OS=Solanum lycopersicum OX=4081 GN=LAT56 PE=2 SV=2 |
| P27762 | 2.25e-28 | 62 | 293 | 67 | 313 | Pectate lyase 4 OS=Ambrosia artemisiifolia OX=4212 PE=1 SV=1 |
| Q9C8G4 | 5.16e-28 | 62 | 299 | 54 | 292 | Probable pectate lyase 4 OS=Arabidopsis thaliana OX=3702 GN=At1g30350 PE=2 SV=1 |
| P40972 | 1.09e-27 | 69 | 293 | 71 | 311 | Pectate lyase OS=Nicotiana tabacum OX=4097 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
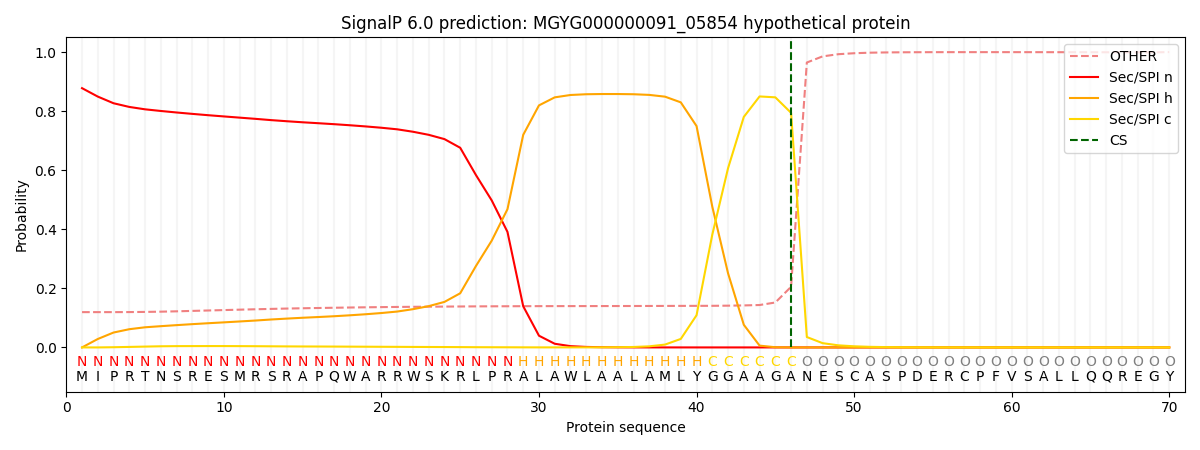
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.123880 | 0.871695 | 0.003073 | 0.000752 | 0.000313 | 0.000285 |
