You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000095_00967
You are here: Home > Sequence: MGYG000000095_00967
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp902363085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp902363085 | |||||||||||
| CAZyme ID | MGYG000000095_00967 | |||||||||||
| CAZy Family | GH66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 281948; End: 283156 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH66 | 66 | 298 | 9.1e-40 | 0.44964028776978415 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14745 | GH66 | 1.28e-64 | 58 | 322 | 43 | 323 | Glycoside Hydrolase Family 66. Glycoside Hydrolase Family 66 contains proteins characterized as cycloisomaltooligosaccharide glucanotransferase (CITase) and dextranases from a variety of bacteria. CITase cyclizes part of a (1-6)-alpha-D-glucan (dextrans) chain by formation of a (1-6)-alpha-D-glucosidic bond. Dextranases catalyze the endohydrolysis of (1-6)-alpha-D-glucosidic linkages in dextran. Some members contain Carbohydrate Binding Module 35 (CBM35) domains, either C-terminal or inserted in the domain or both. |
| pfam13199 | Glyco_hydro_66 | 5.41e-57 | 63 | 400 | 147 | 557 | Glycosyl hydrolase family 66. This family is a set of glycosyl hydrolase enzymes including cycloisomaltooligosaccharide glucanotransferase (EC:2.4.1.-) and dextranase (EC:3.2.1.11) activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AJY77719.1 | 1.40e-86 | 58 | 402 | 150 | 566 |
| QNK59981.1 | 1.16e-84 | 58 | 402 | 172 | 588 |
| AHV97051.1 | 2.79e-84 | 58 | 402 | 153 | 569 |
| AKG37651.1 | 7.22e-84 | 58 | 402 | 150 | 566 |
| QWU13358.1 | 7.22e-84 | 58 | 401 | 150 | 565 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WNL_A | 7.79e-09 | 298 | 401 | 596 | 702 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNM_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNP_A | 7.79e-09 | 298 | 401 | 596 | 702 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNP_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNK_A | 7.84e-09 | 298 | 401 | 615 | 721 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 5X7G_A | 3.19e-08 | 117 | 251 | 258 | 396 | CrystalStructure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase [Paenibacillus sp. 598K],5X7H_A Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose [Paenibacillus sp. 598K] |
| 3VMN_A | 2.04e-06 | 127 | 401 | 265 | 635 | Crystalstructure of dextranase from Streptococcus mutans [Streptococcus mutans],3VMO_A Crystal structure of dextranase from Streptococcus mutans in complex with isomaltotriose [Streptococcus mutans],3VMP_A Crystal structure of dextranase from Streptococcus mutans in complex with 4,5-epoxypentyl alpha-D-glucopyranoside [Streptococcus mutans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94286 | 2.71e-08 | 298 | 402 | 632 | 739 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
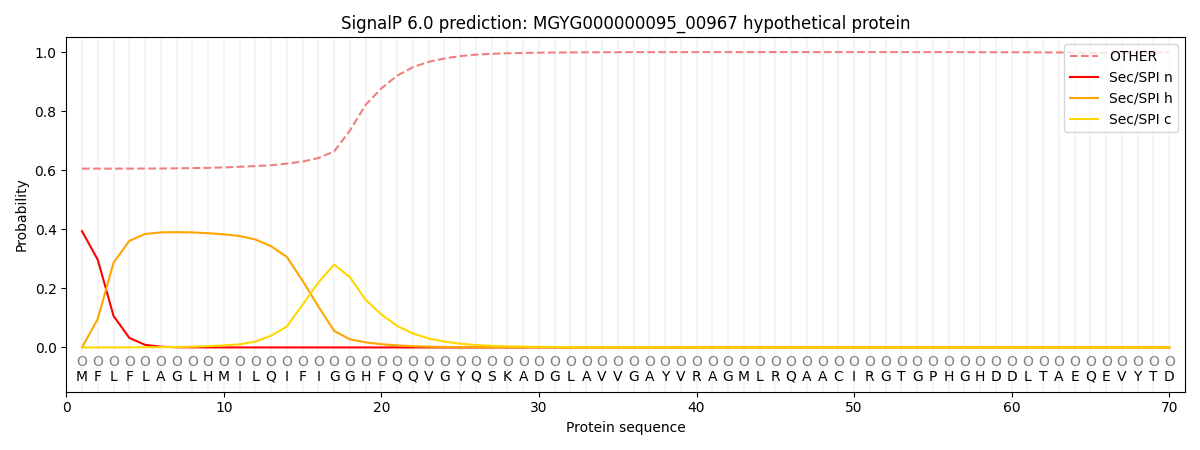
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.625328 | 0.370946 | 0.001659 | 0.000510 | 0.000395 | 0.001161 |
