You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000095_01275
You are here: Home > Sequence: MGYG000000095_01275
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
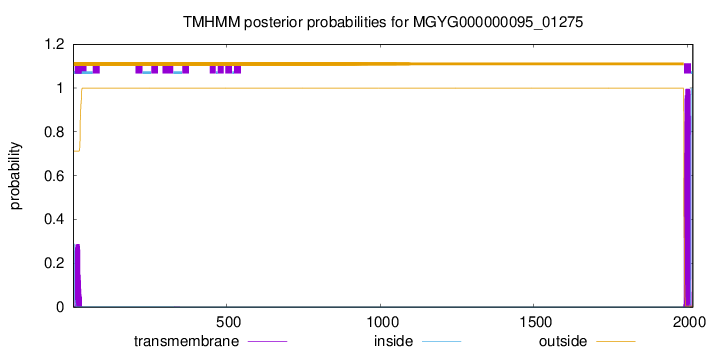
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp902363085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp902363085 | |||||||||||
| CAZyme ID | MGYG000000095_01275 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 184536; End: 190592 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM38 | 1609 | 1732 | 2.1e-18 | 0.9767441860465116 |
| CBM32 | 47 | 157 | 1.2e-17 | 0.8467741935483871 |
| CBM38 | 1459 | 1589 | 2.7e-17 | 0.9534883720930233 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 8.08e-13 | 47 | 168 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02449 | Glyco_hydro_42 | 3.65e-10 | 249 | 442 | 41 | 233 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| cd00057 | FA58C | 6.47e-07 | 40 | 163 | 6 | 136 | Substituted updates: Jan 31, 2002 |
| PHA03418 | PHA03418 | 0.001 | 1930 | 1998 | 53 | 128 | hypothetical E4 protein; Provisional |
| COG4932 | COG4932 | 0.001 | 1957 | 2018 | 1470 | 1531 | Uncharacterized surface anchored protein [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBI31181.1 | 0.0 | 3 | 1895 | 51 | 1751 |
| QJD86795.1 | 0.0 | 40 | 1895 | 52 | 1716 |
| QTH42816.1 | 3.18e-18 | 1385 | 1611 | 140 | 355 |
| AGA28800.1 | 1.77e-13 | 1433 | 1608 | 18 | 191 |
| QEV29298.1 | 1.57e-11 | 1371 | 1604 | 313 | 534 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5FJ41 | 3.79e-06 | 252 | 396 | 55 | 198 | Beta-galactosidase LacZ OS=Lactobacillus acidophilus (strain ATCC 700396 / NCK56 / N2 / NCFM) OX=272621 GN=lacZ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
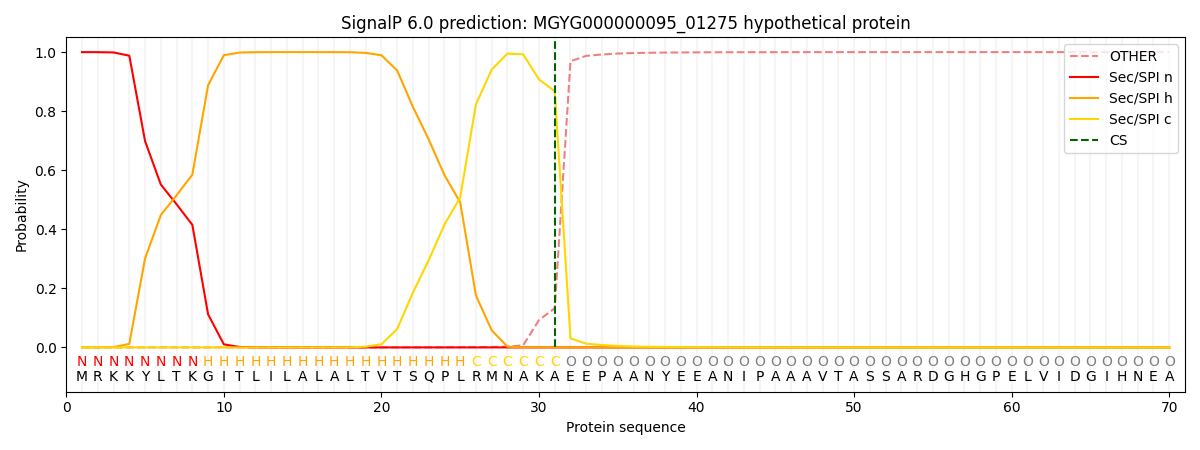
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000390 | 0.998649 | 0.000405 | 0.000204 | 0.000176 | 0.000160 |