You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000095_02128
You are here: Home > Sequence: MGYG000000095_02128
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp902363085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp902363085 | |||||||||||
| CAZyme ID | MGYG000000095_02128 | |||||||||||
| CAZy Family | GH98 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 326540; End: 329149 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH98 | 32 | 329 | 3.4e-89 | 0.9938837920489296 |
| CBM47 | 610 | 730 | 2.7e-35 | 0.9921875 |
| CBM47 | 745 | 864 | 1.3e-34 | 0.9921875 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08306 | Glyco_hydro_98M | 4.74e-97 | 29 | 328 | 2 | 328 | Glycosyl hydrolase family 98. This domain is the putative catalytic domain of glycosyl hydrolase family 98 proteins. |
| pfam08307 | Glyco_hydro_98C | 1.05e-86 | 331 | 584 | 1 | 268 | Glycosyl hydrolase family 98 C-terminal domain. This putative domain is found at the C-terminus of glycosyl hydrolase family 98 proteins. This domain is not expected to form part of the catalytic activity. |
| smart00607 | FTP | 1.96e-19 | 603 | 733 | 3 | 145 | eel-Fucolectin Tachylectin-4 Pentaxrin-1 Domain. |
| smart00607 | FTP | 4.73e-17 | 737 | 868 | 2 | 146 | eel-Fucolectin Tachylectin-4 Pentaxrin-1 Domain. |
| pfam00754 | F5_F8_type_C | 3.69e-13 | 744 | 864 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANO37934.1 | 0.0 | 2 | 869 | 4 | 895 |
| AOG59126.1 | 0.0 | 2 | 869 | 4 | 895 |
| VFH40922.1 | 0.0 | 2 | 869 | 4 | 895 |
| VFI11899.1 | 0.0 | 2 | 869 | 4 | 895 |
| AVD76032.1 | 0.0 | 2 | 869 | 4 | 895 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2WMH_A | 6.25e-262 | 32 | 583 | 27 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 in complex with the H- disaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
| 2WMF_A | 3.57e-261 | 32 | 583 | 27 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in its native form. [Streptococcus pneumoniae TIGR4] |
| 2WMG_A | 1.02e-260 | 32 | 583 | 27 | 581 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae TIGR4 (Sp4GH98) in complex with the LewisY pentasaccharide blood group antigen. [Streptococcus pneumoniae TIGR4] |
| 2WMI_A | 1.94e-115 | 21 | 584 | 14 | 605 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 in complex with the A-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71],2WMJ_A Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71],2WMJ_B Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the B-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71] |
| 2WMI_B | 3.82e-115 | 21 | 584 | 14 | 605 | Crystalstructure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 in complex with the A-trisaccharide blood group antigen. [Streptococcus pneumoniae SP3-BS71] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6RUF5 | 3.09e-115 | 35 | 583 | 221 | 797 | Blood-group-substance endo-1,4-beta-galactosidase OS=Clostridium perfringens OX=1502 GN=eabC PE=1 SV=1 |
| Q9I928 | 1.65e-20 | 597 | 735 | 27 | 175 | Fucolectin-4 OS=Anguilla japonica OX=7937 PE=2 SV=1 |
| Q9I931 | 4.06e-20 | 599 | 735 | 27 | 174 | Fucolectin-1 OS=Anguilla japonica OX=7937 PE=2 SV=1 |
| Q9I927 | 1.58e-18 | 597 | 735 | 36 | 185 | Fucolectin-5 OS=Anguilla japonica OX=7937 PE=2 SV=1 |
| Q9I929 | 1.48e-17 | 599 | 733 | 29 | 173 | Fucolectin-3 OS=Anguilla japonica OX=7937 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
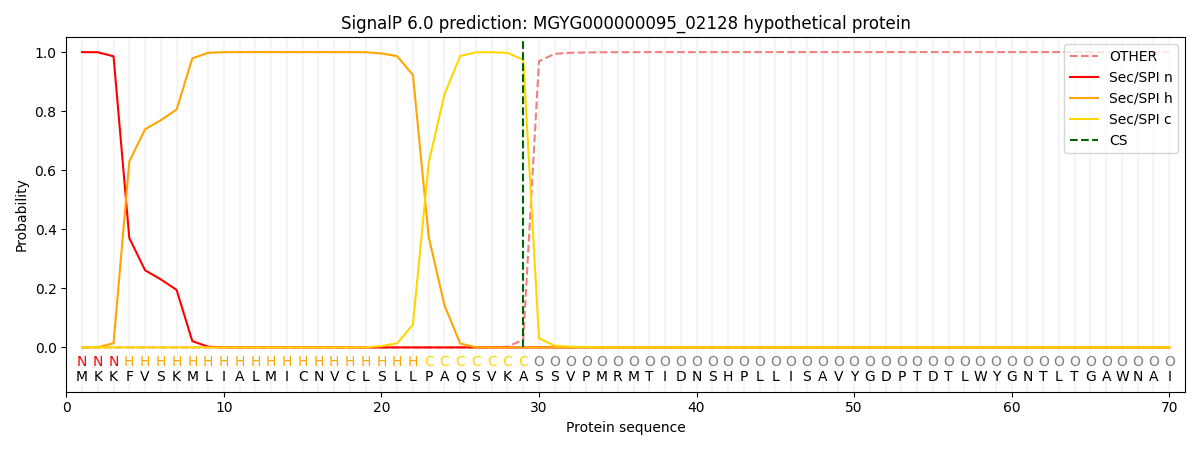
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000285 | 0.999000 | 0.000198 | 0.000183 | 0.000154 | 0.000141 |
