You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000095_02405
You are here: Home > Sequence: MGYG000000095_02405
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp902363085 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp902363085 | |||||||||||
| CAZyme ID | MGYG000000095_02405 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2086; End: 4332 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 39 | 449 | 3.5e-92 | 0.9933333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 2.54e-49 | 8 | 450 | 5 | 426 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam02057 | Glyco_hydro_59 | 1.07e-25 | 67 | 321 | 19 | 251 | Glycosyl hydrolase family 59. |
| pfam14587 | Glyco_hydr_30_2 | 3.41e-18 | 39 | 322 | 4 | 354 | O-Glycosyl hydrolase family 30. |
| pfam02055 | Glyco_hydro_30 | 2.16e-15 | 133 | 358 | 95 | 348 | Glycosyl hydrolase family 30 TIM-barrel domain. |
| pfam17189 | Glyco_hydro_30C | 3.68e-04 | 361 | 450 | 1 | 63 | Glycosyl hydrolase family 30 beta sandwich domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS14928.1 | 5.12e-146 | 37 | 598 | 33 | 597 |
| ASS68692.1 | 2.33e-141 | 5 | 598 | 8 | 599 |
| QUH30730.1 | 2.56e-134 | 39 | 601 | 34 | 593 |
| QOV18445.1 | 2.30e-109 | 39 | 452 | 5 | 424 |
| QGQ97720.1 | 6.16e-100 | 6 | 457 | 23 | 487 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5CXP_A | 3.10e-44 | 48 | 450 | 15 | 385 | X-raycrystallographic protein structure of the glycoside hydrolase family 30 subfamily 8 xylanase, Xyn30A, from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
| 4FMV_A | 4.85e-44 | 38 | 452 | 4 | 386 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
| 6IUJ_A | 2.76e-42 | 16 | 450 | 1 | 469 | ChainA, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612],6IUJ_B Chain B, GH30 Xylanase B [Talaromyces cellulolyticus CF-2612] |
| 6M5Z_A | 2.71e-40 | 39 | 450 | 25 | 451 | ChainA, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612],6M5Z_B Chain B, GH30 Xylanase C [Talaromyces cellulolyticus CF-2612] |
| 6KRL_A | 8.24e-40 | 38 | 450 | 94 | 540 | Crystalstructure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris [Saccharomyces uvarum],6KRN_A Crystal structure of GH30 xylanase B from Talaromyces cellulolyticus expressed by Pichia pastoris in complex with aldotriuronic acid [Saccharomyces uvarum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2Q1N4 | 2.38e-36 | 41 | 449 | 31 | 468 | GH30 family xylanase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Xyn30A PE=1 SV=1 |
| Q6YK37 | 3.89e-32 | 1 | 450 | 2 | 419 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
| Q45070 | 1.72e-31 | 84 | 450 | 76 | 418 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| P54818 | 9.10e-13 | 48 | 289 | 53 | 278 | Galactocerebrosidase OS=Mus musculus OX=10090 GN=Galc PE=1 SV=2 |
| O16580 | 1.58e-12 | 48 | 397 | 96 | 484 | Putative glucosylceramidase 1 OS=Caenorhabditis elegans OX=6239 GN=gba-1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
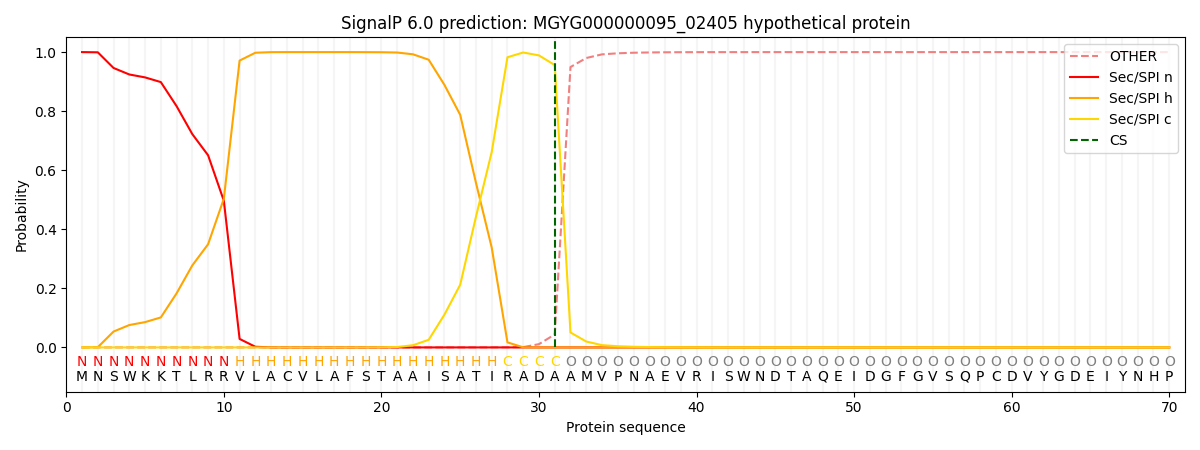
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000262 | 0.999042 | 0.000170 | 0.000192 | 0.000160 | 0.000142 |
