You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000096_01381
You are here: Home > Sequence: MGYG000000096_01381
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
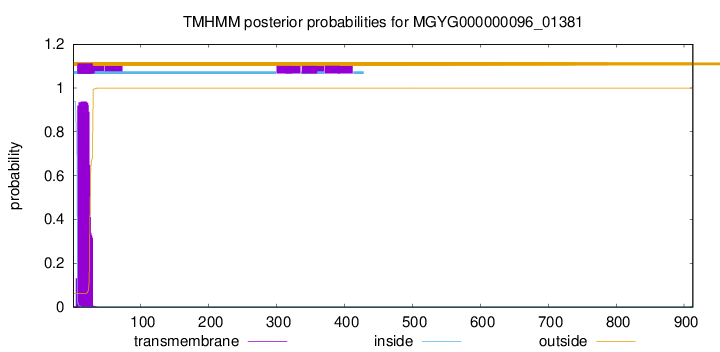
TMHMM annotations
Basic Information help
| Species | Leuconostoc pseudomesenteroides | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Leuconostoc; Leuconostoc pseudomesenteroides | |||||||||||
| CAZyme ID | MGYG000000096_01381 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 93837; End: 96578 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 759 | 908 | 7.7e-34 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1705 | FlgJ | 2.84e-39 | 726 | 913 | 20 | 188 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| PRK05684 | flgJ | 1.04e-27 | 771 | 907 | 174 | 300 | flagellar assembly peptidoglycan hydrolase FlgJ. |
| PRK08581 | PRK08581 | 1.02e-25 | 688 | 913 | 272 | 472 | amidase domain-containing protein. |
| TIGR02541 | flagell_FlgJ | 4.81e-24 | 754 | 905 | 153 | 291 | flagellar rod assembly protein/muramidase FlgJ. The N-terminal region of this protein acts directly in flagellar rod assembly. The C-terminal region is a flagellum-specific muramidase (peptidoglycan hydrolase) required for formation of the outer membrane L ring. |
| COG5492 | YjdB | 3.30e-23 | 315 | 591 | 1 | 323 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATC61482.1 | 1.03e-177 | 1 | 913 | 1 | 974 |
| QIW57581.1 | 9.14e-150 | 1 | 913 | 1 | 975 |
| QEA46519.1 | 2.66e-145 | 3 | 913 | 2 | 850 |
| QEA63209.1 | 2.66e-145 | 3 | 913 | 2 | 850 |
| QEA54932.1 | 3.74e-145 | 3 | 913 | 2 | 850 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3LAT_A | 3.94e-29 | 26 | 224 | 17 | 204 | Crystalstructure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis],3LAT_B Crystal structure of Staphylococcus peptidoglycan hydrolase AmiE [Staphylococcus epidermidis] |
| 4KNK_A | 5.40e-25 | 26 | 224 | 26 | 213 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNK_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_A Chain A, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_B Chain B, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_C Chain C, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325],4KNL_D Chain D, Bifunctional autolysin [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 3FI7_A | 6.43e-23 | 747 | 913 | 28 | 183 | CrystalStructure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain [Listeria monocytogenes EGD-e] |
| 5DN5_A | 7.05e-17 | 773 | 905 | 27 | 146 | Structureof a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_B Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_C Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 5DN4_A | 1.02e-16 | 773 | 905 | 27 | 146 | Structureof the glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CPQ1 | 1.38e-26 | 26 | 257 | 319 | 536 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q5HQB9 | 1.38e-26 | 26 | 257 | 319 | 536 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| O33635 | 1.38e-26 | 26 | 257 | 319 | 536 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q6GAG0 | 2.83e-23 | 26 | 252 | 222 | 438 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q2FZK7 | 9.95e-22 | 26 | 224 | 222 | 409 | Bifunctional autolysin OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
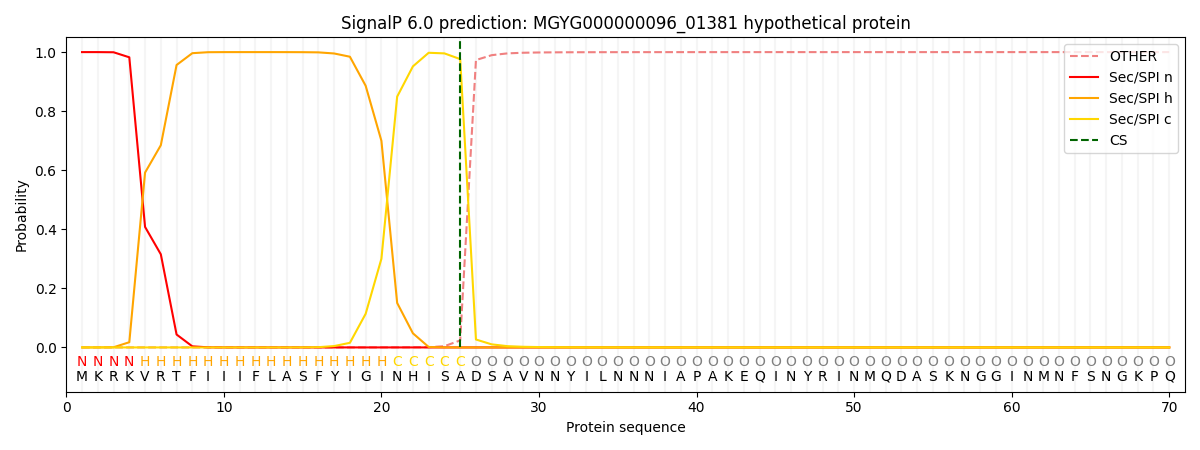
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000344 | 0.998917 | 0.000242 | 0.000162 | 0.000161 | 0.000152 |