You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000098_00792
You are here: Home > Sequence: MGYG000000098_00792
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000098_00792 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61949; End: 63289 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 317 | 440 | 2.7e-19 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.07e-16 | 313 | 440 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam08522 | DUF1735 | 3.83e-12 | 212 | 285 | 40 | 117 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| cd00057 | FA58C | 3.97e-08 | 303 | 431 | 3 | 134 | Substituted updates: Jan 31, 2002 |
| pfam08522 | DUF1735 | 2.25e-07 | 35 | 159 | 1 | 120 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| smart00231 | FA58C | 4.44e-05 | 316 | 444 | 16 | 138 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT72760.1 | 1.94e-153 | 2 | 445 | 3 | 448 |
| BCA52367.1 | 1.31e-145 | 9 | 445 | 8 | 446 |
| QUT73146.1 | 1.31e-145 | 9 | 445 | 8 | 446 |
| QUT40999.1 | 1.06e-144 | 9 | 445 | 8 | 446 |
| QUT88787.1 | 6.61e-141 | 9 | 445 | 7 | 446 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3F2Z_A | 1.05e-20 | 304 | 444 | 2 | 147 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
| 3GGL_A | 1.35e-20 | 304 | 444 | 12 | 157 | X-RayStructure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. [Bacteroides thetaiotaomicron],6OE2_A X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. Re-refinement of 3GGL with correct metal Mn replacing Zn. New metal confirmed with PIXE analysis of original sample. [Bacteroides thetaiotaomicron] |
| 2KD7_A | 1.44e-20 | 303 | 444 | 1 | 147 | ChainA, Putative chitobiase [Bacteroides thetaiotaomicron VPI-5482] |
| 2J7M_A | 6.32e-11 | 314 | 444 | 13 | 149 | Characterizationof a Family 32 CBM [Clostridium perfringens] |
| 2J1A_A | 6.46e-11 | 314 | 444 | 14 | 150 | Structureof CBM32 from Clostridium perfringens beta-N- acetylhexosaminidase GH84C in complex with galactose [Clostridium perfringens ATCC 13124],2J1E_A High Resolution Crystal Structure of CBM32 from a N-acetyl-beta- hexosaminidase in complex with lacNAc [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 8.59e-09 | 314 | 444 | 631 | 767 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 8.59e-09 | 314 | 444 | 631 | 767 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
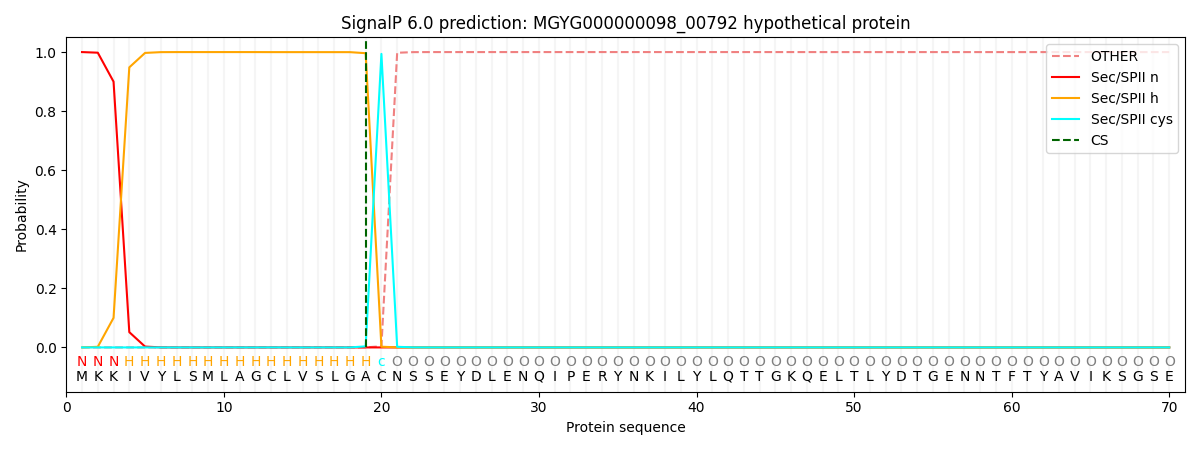
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000063 | 0.000000 | 0.000000 | 0.000000 |
