You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000098_00795
You are here: Home > Sequence: MGYG000000098_00795
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000098_00795 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66635; End: 69442 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 23 | 718 | 1.2e-66 | 0.7061170212765957 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 1.13e-21 | 29 | 533 | 13 | 500 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 3.75e-15 | 30 | 343 | 14 | 296 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 7.50e-14 | 31 | 450 | 44 | 455 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02837 | Glyco_hydro_2_N | 1.10e-09 | 30 | 221 | 3 | 169 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| pfam00703 | Glyco_hydro_2 | 1.30e-07 | 225 | 323 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75482.1 | 0.0 | 11 | 931 | 7 | 932 |
| QRO25183.1 | 0.0 | 15 | 935 | 13 | 935 |
| QEL03576.1 | 0.0 | 6 | 918 | 3 | 929 |
| SDS50045.1 | 0.0 | 13 | 930 | 14 | 944 |
| AQS94810.1 | 0.0 | 13 | 930 | 14 | 943 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D8K_A | 3.14e-18 | 53 | 474 | 36 | 443 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 3GM8_A | 4.85e-16 | 118 | 478 | 66 | 428 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
| 6D4O_A | 1.47e-12 | 17 | 454 | 4 | 437 | Eubacteriumeligens beta-glucuronidase bound to an amoxapine-glucuronide conjugate [Lachnospira eligens] |
| 6BO6_A | 1.93e-12 | 17 | 454 | 1 | 434 | Eubacteriumeligens beta-glucuronidase bound to UNC4917 glucuronic acid conjugate [[Eubacterium] eligens ATCC 27750] |
| 6BJW_A | 2.00e-12 | 17 | 454 | 25 | 458 | ChainA, Glycoside Hydrolase Family 2 candidate b-glucuronidase [[Eubacterium] eligens ATCC 27750] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q04F24 | 1.18e-16 | 30 | 466 | 49 | 475 | Beta-galactosidase OS=Oenococcus oeni (strain ATCC BAA-331 / PSU-1) OX=203123 GN=lacZ PE=3 SV=1 |
| Q6LL68 | 1.81e-15 | 67 | 493 | 62 | 501 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| Q8D4H3 | 2.77e-14 | 56 | 493 | 45 | 503 | Beta-galactosidase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=lacZ PE=3 SV=2 |
| Q7MG04 | 6.28e-14 | 56 | 493 | 45 | 502 | Beta-galactosidase OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=lacZ PE=3 SV=1 |
| P81650 | 1.09e-13 | 20 | 465 | 39 | 476 | Beta-galactosidase OS=Pseudoalteromonas haloplanktis OX=228 GN=lacZ PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
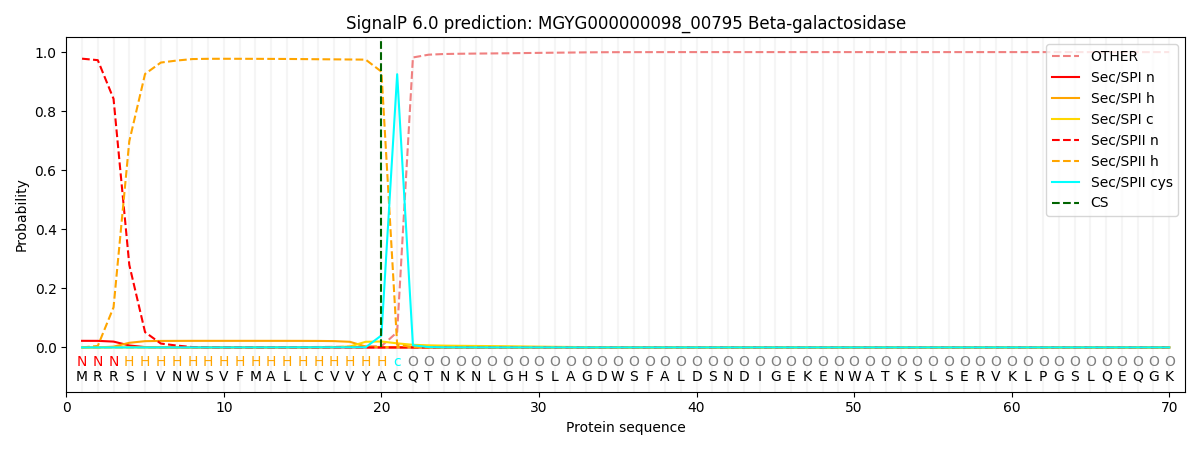
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000074 | 0.021734 | 0.978199 | 0.000008 | 0.000012 | 0.000010 |
