You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000098_00820
You are here: Home > Sequence: MGYG000000098_00820
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000098_00820 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 102078; End: 104327 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 457 | 728 | 2.8e-36 | 0.7415384615384616 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.16e-19 | 427 | 668 | 118 | 399 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 1.90e-13 | 459 | 733 | 46 | 305 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03010 | PLN03010 | 1.39e-10 | 504 | 678 | 167 | 318 | polygalacturonase |
| PLN03003 | PLN03003 | 3.89e-07 | 459 | 678 | 108 | 299 | Probable polygalacturonase At3g15720 |
| PLN02793 | PLN02793 | 5.71e-07 | 459 | 678 | 137 | 338 | Probable polygalacturonase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL02017.1 | 9.96e-260 | 1 | 738 | 1 | 732 |
| QRP58902.1 | 1.10e-220 | 22 | 735 | 44 | 760 |
| QQA09745.1 | 1.10e-220 | 22 | 735 | 44 | 760 |
| QQT79214.1 | 1.10e-220 | 22 | 735 | 44 | 760 |
| ASW16403.1 | 1.10e-220 | 22 | 735 | 44 | 760 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A2QW66 | 2.59e-11 | 453 | 708 | 124 | 378 | Probable exopolygalacturonase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pgxA PE=3 SV=1 |
| Q27UB2 | 2.59e-11 | 453 | 708 | 124 | 378 | Exopolygalacturonase A OS=Aspergillus niger OX=5061 GN=pgxA PE=3 SV=1 |
| P49062 | 1.78e-10 | 459 | 695 | 131 | 352 | Exopolygalacturonase clone GBGE184 OS=Arabidopsis thaliana OX=3702 GN=PGA3 PE=2 SV=1 |
| B0YAA4 | 1.87e-09 | 463 | 699 | 141 | 374 | Probable exopolygalacturonase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=pgxB PE=3 SV=2 |
| Q4WBK6 | 1.87e-09 | 463 | 699 | 141 | 374 | Probable exopolygalacturonase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pgxB PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
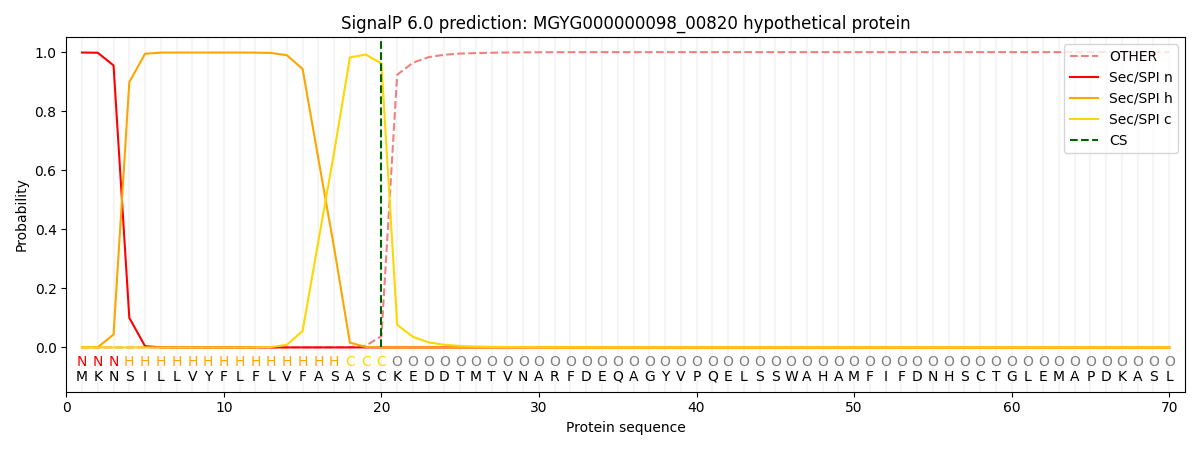
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000483 | 0.996656 | 0.002281 | 0.000202 | 0.000180 | 0.000169 |
