You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000098_01160
You are here: Home > Sequence: MGYG000000098_01160
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000098_01160 | |||||||||||
| CAZy Family | GH38 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 106197; End: 108758 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH38 | 44 | 190 | 1.7e-29 | 0.5315985130111525 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0383 | AMS1 | 6.40e-19 | 104 | 818 | 286 | 917 | Alpha-mannosidase [Carbohydrate transport and metabolism]. |
| PRK09819 | PRK09819 | 6.58e-12 | 74 | 837 | 66 | 862 | mannosylglycerate hydrolase. |
| pfam01074 | Glyco_hydro_38 | 1.63e-11 | 74 | 188 | 60 | 172 | Glycosyl hydrolases family 38 N-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. |
| pfam07748 | Glyco_hydro_38C | 3.98e-10 | 493 | 694 | 2 | 184 | Glycosyl hydrolases family 38 C-terminal domain. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. |
| smart00872 | Alpha-mann_mid | 1.57e-09 | 288 | 343 | 8 | 62 | Alpha mannosidase, middle domain. Members of this entry belong to the glycosyl hydrolase family 38, This domain, which is found in the central region adopts a structure consisting of three alpha helices, in an immunoglobulin/albumin-binding domain-like fold. The domain is predominantly found in the enzyme alpha-mannosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUR41760.1 | 0.0 | 1 | 852 | 1 | 853 |
| QQA07855.1 | 0.0 | 6 | 853 | 7 | 849 |
| QRM98564.1 | 0.0 | 1 | 852 | 1 | 853 |
| QNL41054.1 | 0.0 | 1 | 853 | 1 | 855 |
| QUT27309.1 | 0.0 | 1 | 852 | 1 | 853 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q54K67 | 5.92e-10 | 49 | 848 | 291 | 1082 | Alpha-mannosidase G OS=Dictyostelium discoideum OX=44689 GN=manG PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
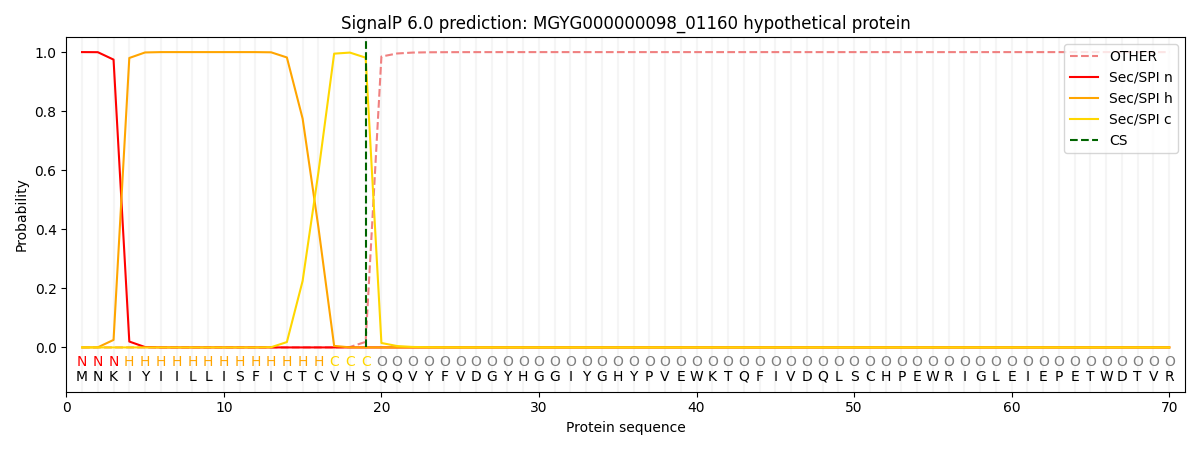
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000230 | 0.999178 | 0.000173 | 0.000138 | 0.000137 | 0.000134 |
