You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000098_01295
You are here: Home > Sequence: MGYG000000098_01295
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000098_01295 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 97991; End: 99184 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam09594 | GT87 | 6.93e-26 | 66 | 291 | 1 | 235 | Glycosyltransferase family 87. The enzymes in this family are glycosyltransferases. PimE is involved in phosphatidylinositol mannoside (PIM) synthesis, a major class of glycolipids in all mycobacteria. PimE is a polyprenol-phosphate-mannose-dependent mannosyltransferase that transfers the fifth mannose of PIM. The family also includes alpha(1-->3) arabinofuranosyltransferase, invloved in the synthesis of of mycobacterial arabinogalactan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJE30394.1 | 9.75e-140 | 6 | 394 | 8 | 399 |
| QUR48935.1 | 9.75e-140 | 6 | 394 | 8 | 399 |
| QIX65609.1 | 9.75e-140 | 6 | 394 | 8 | 399 |
| QUT20372.1 | 1.38e-139 | 6 | 394 | 8 | 399 |
| QUT52835.1 | 5.52e-139 | 6 | 394 | 8 | 399 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
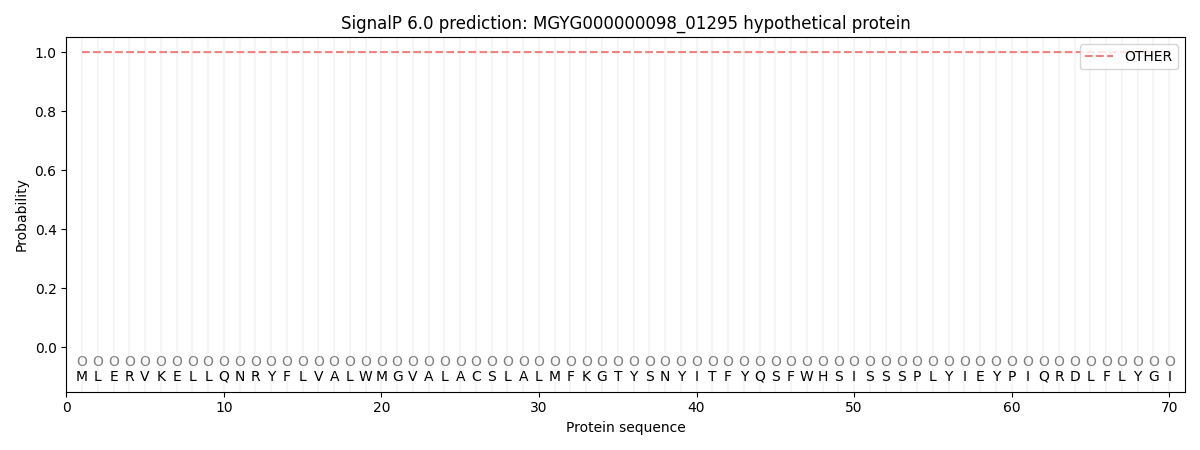
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000025 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
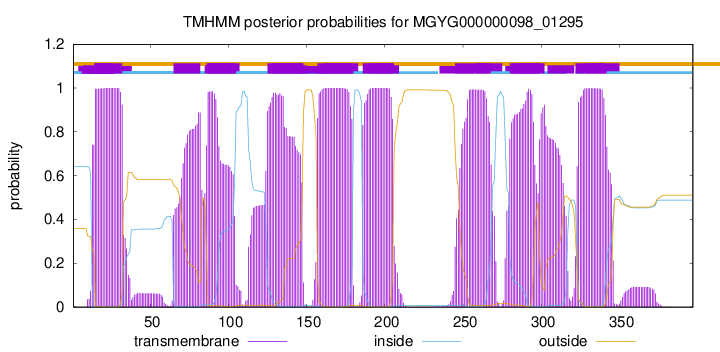
| start | end |
|---|---|
| 15 | 32 |
| 65 | 82 |
| 87 | 104 |
| 125 | 147 |
| 157 | 179 |
| 186 | 205 |
| 245 | 267 |
| 280 | 302 |
| 322 | 341 |
