You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000098_01323
You are here: Home > Sequence: MGYG000000098_01323
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000098_01323 | |||||||||||
| CAZy Family | CBM62 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 126123; End: 128096 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM62 | 529 | 589 | 8.3e-18 | 0.4580152671755725 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 0.008 | 540 | 619 | 7 | 87 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd09361 | LIM1_Enigma_like | 0.009 | 502 | 529 | 18 | 49 | The first LIM domain of Enigma-like family. The first LIM domain of Enigma-like family: The Enigma LIM domain family is comprised of three members: Enigma, ENH, and Cypher (mouse)/ZASP (human). These subfamily members contain a single PDZ domain at the N-terminus and three LIM domains at the C-terminus. Enigma was initially characterized in humans and is expressed in multiple tissues, such as skeletal muscle, heart, bone, and brain. The third LIM domain specifically interacts with the insulin receptor and the second LIM domain interacts with the receptor tyrosine kinase Ret and the adaptor protein APS. Thus Enigma is implicated in signal transduction processes, such as mitogenic activity, insulin related actin organization, and glucose metabolism. The second member, ENH protein, was first identified in rat brain. It has been shown that ENH interacts with protein kinase D1 (PKD1) via its LIM domains and forms a complex with PKD1 and the alpha1C subunit of cardiac L-type voltage-gated calcium channel in rat neonatal cardiomyocytes. The N-terminal PDZ domain interacts with alpha-actinin at the Z-line. ZASP/Cypher is required for maintenance of Z-line structure during muscle contraction, but not required for Z-line assembly. In heart, Cypher/ZASP plays a structural role through its interaction with cytoskeletal Z-line proteins. In addition, there is increasing evidence that Cypher/ZASP also performs signaling functions. Studies reveal that Cypher/ZASP interacts with and directs PKC to the Z-line, where PKC phosphorylates downstream signaling targets. LIM domains are 50-60 amino acids in size and share two characteristic zinc finger motifs. The two zinc fingers contain eight conserved residues, mostly cysteines and histidines, which coordinately bond to two zinc atoms. LIM domains function as adaptors or scaffolds to support the assembly of multimeric protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74079.1 | 1.88e-255 | 23 | 657 | 18 | 648 |
| ABR45148.1 | 3.63e-178 | 16 | 657 | 6 | 660 |
| QCY54736.1 | 3.63e-178 | 16 | 657 | 6 | 660 |
| QRO14748.1 | 3.63e-178 | 16 | 657 | 6 | 660 |
| QUT52565.1 | 3.63e-178 | 16 | 657 | 6 | 660 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2YB7_A | 1.34e-09 | 528 | 600 | 7 | 80 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YB7_B Chain B, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFU_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YFZ_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],2YG0_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5G56_A | 1.59e-08 | 528 | 600 | 708 | 781 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
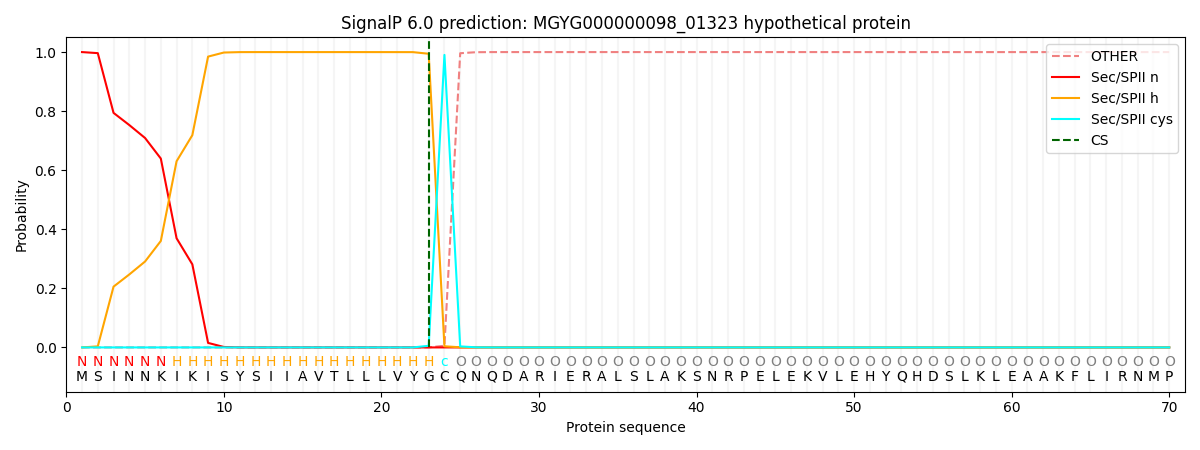
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000050 | 0.000000 | 0.000000 | 0.000000 |
