You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000098_02421
You are here: Home > Sequence: MGYG000000098_02421
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
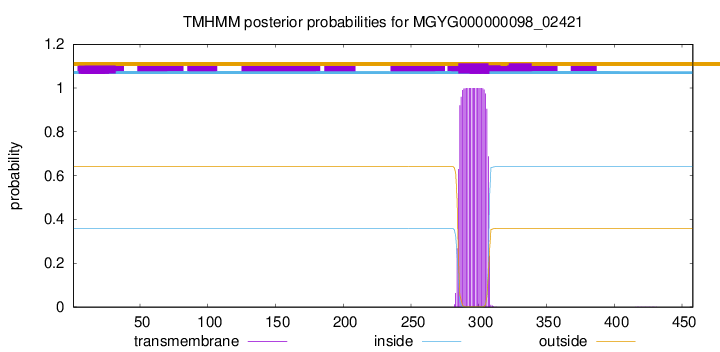
TMHMM annotations
Basic Information help
| Species | Bacteroides bouchesdurhonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides bouchesdurhonensis | |||||||||||
| CAZyme ID | MGYG000000098_02421 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Integration host factor subunit alpha | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 72314; End: 73690 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00216 | Bac_DNA_binding | 2.61e-17 | 6 | 93 | 1 | 88 | Bacterial DNA-binding protein. |
| smart00411 | BHL | 2.52e-12 | 5 | 94 | 1 | 90 | bacterial (prokaryotic) histone like domain. |
| COG0776 | HimA | 6.14e-12 | 5 | 96 | 2 | 93 | Bacterial nucleoid DNA-binding protein [Replication, recombination and repair]. |
| cd13835 | IHF_A | 1.76e-11 | 6 | 93 | 1 | 88 | Alpha subunit of integration host factor (IHFA). This subfamily consists of the alpha subunit of integration host factor (IHF) and IHF-like domains. IHF is a nucleoid-associated protein (NAP) that binds and sharply bends many DNA targets in a sequence specific manner. It is a heterodimeric protein composed of two highly homologous subunits IHFA (IHF-alpha) and IHFB (IHF-beta). It is known to act as a transcription factor at many gene regulatory regions in E. coli. IHF is an essential cofactor in phage lambda site-specific recombination, having an architectural role during assembly of specialized nucleoprotein structures (snups). IHF is also involved in formation as well as maintenance of bacterial biofilms since it is found in complex with extracellular DNA (eDNA) within the extracellular polymeric substances (EPS) matrix of many biofilms. |
| cd13832 | IHF | 2.83e-11 | 9 | 91 | 3 | 85 | Integration host factor (IHF) and similar proteins. This subfamily includes integration host factor (IHF) and IHF-like domains. IHF is a nucleoid-associated protein (NAP) that binds and sharply bends many DNA targets in a sequence specific manner. It is a heterodimeric protein composed of two highly homologous subunits IHFA (IHF-alpha) and IHFB (IHF-beta). It is known to act as a transcription factor at many gene regulatory regions in E. coli. IHF is an essential cofactor in phage lambda site-specific recombination, having an architectural role during assembly of specialized nucleoprotein structures (snups). IHF is also involved in formation as well as maintenance of bacterial biofilms since it is found in complex with extracellular DNA (eDNA) within the extracellular polymeric substances (EPS) matrix of many biofilms. This subfamily also includes the protein Hbb from tick-borne spirochete Borrelia burgdorferi, responsible for causing Lyme disease in humans. Hbb, a homodimer, shows DNA sequence preferences that are related, yet distinct from those of IHF. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQR17553.1 | 3.68e-140 | 1 | 457 | 1 | 443 |
| ANU57570.1 | 3.68e-140 | 1 | 457 | 1 | 443 |
| BCA50162.1 | 7.92e-137 | 1 | 457 | 1 | 486 |
| QNL37743.1 | 1.23e-134 | 1 | 457 | 1 | 470 |
| QUT81732.1 | 2.46e-134 | 1 | 457 | 1 | 470 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J0N_I | 1.02e-06 | 5 | 93 | 2 | 90 | ChainI, Integration host factor subunit alpha [Escherichia coli],5J0N_K Chain K, Integration host factor subunit alpha [Escherichia coli] |
| 1IHF_A | 1.09e-06 | 5 | 93 | 3 | 91 | INTEGRATIONHOST FACTOR/DNA COMPLEX [Escherichia coli],1OUZ_A Chain A, Integration Host Factor Alpha-subunit [Escherichia coli],1OWF_A Chain A, Integration Host Factor Alpha-subunit [Escherichia coli],1OWG_A Crystal structure of WT IHF complexed with an altered H' site (T44A) [Escherichia coli],2HT0_A IHF bound to doubly nicked DNA [Escherichia coli],5WFE_K Cas1-Cas2-IHF-DNA holo-complex [Escherichia coli S88] |
| 2IIE_A | 6.82e-06 | 5 | 93 | 47 | 135 | singlechain Integration Host Factor protein (scIHF2) in complex with DNA [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q65TL4 | 1.56e-08 | 5 | 95 | 3 | 93 | Integration host factor subunit alpha OS=Mannheimia succiniciproducens (strain MBEL55E) OX=221988 GN=ihfA PE=3 SV=1 |
| Q9CN18 | 1.39e-07 | 5 | 95 | 3 | 93 | Integration host factor subunit alpha OS=Pasteurella multocida (strain Pm70) OX=272843 GN=ihfA PE=3 SV=1 |
| Q47CN0 | 6.91e-07 | 5 | 97 | 3 | 95 | Integration host factor subunit alpha 2 OS=Dechloromonas aromatica (strain RCB) OX=159087 GN=ihfA2 PE=3 SV=1 |
| B5FDX6 | 1.23e-06 | 5 | 90 | 3 | 88 | Integration host factor subunit alpha OS=Aliivibrio fischeri (strain MJ11) OX=388396 GN=ihfA PE=3 SV=1 |
| B6EN06 | 1.23e-06 | 5 | 90 | 3 | 88 | Integration host factor subunit alpha OS=Aliivibrio salmonicida (strain LFI1238) OX=316275 GN=ihfA PE=3 SV=1 |
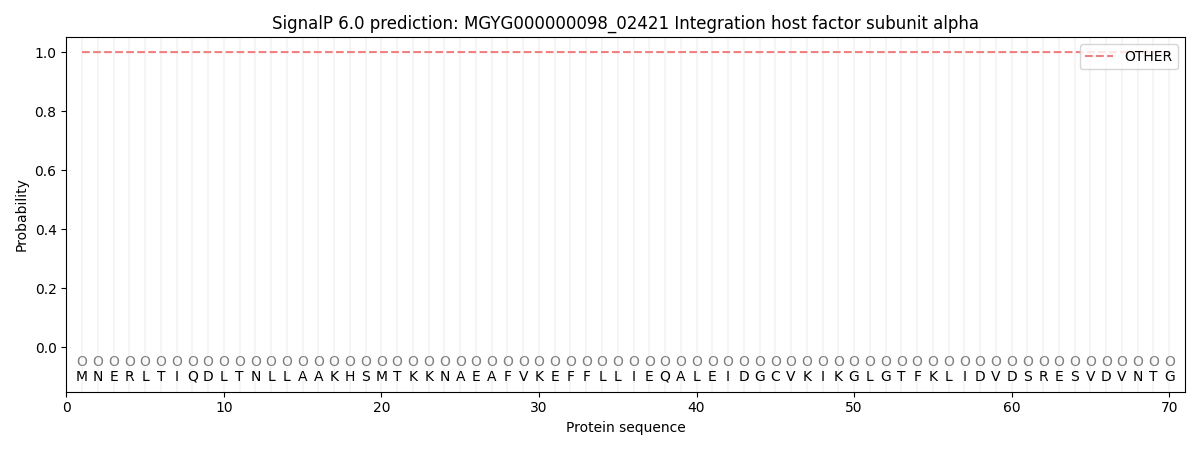
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000067 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |