You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000101_00636
You are here: Home > Sequence: MGYG000000101_00636
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Micrococcus luteus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Micrococcaceae; Micrococcus; Micrococcus luteus | |||||||||||
| CAZyme ID | MGYG000000101_00636 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 340249; End: 341433 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 278 | 391 | 3.5e-17 | 0.837037037037037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16896 | LT_Slt70-like | 6.18e-18 | 268 | 390 | 1 | 143 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd00254 | LT-like | 7.72e-17 | 286 | 392 | 1 | 111 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| PRK06347 | PRK06347 | 1.88e-16 | 41 | 224 | 330 | 527 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 1.19e-14 | 30 | 220 | 394 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| pfam01464 | SLT | 3.57e-13 | 275 | 383 | 1 | 111 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGS21047.1 | 2.76e-267 | 6 | 394 | 1 | 390 |
| QHG59983.1 | 2.76e-267 | 6 | 394 | 1 | 390 |
| QDW17614.1 | 2.82e-260 | 6 | 394 | 1 | 390 |
| SQG47298.1 | 5.63e-259 | 1 | 394 | 1 | 395 |
| ACS30879.1 | 8.62e-255 | 6 | 394 | 1 | 390 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q7A7E0 | 1.32e-13 | 34 | 221 | 19 | 202 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain N315) OX=158879 GN=sle1 PE=1 SV=1 |
| Q5HIL2 | 1.32e-13 | 34 | 221 | 19 | 202 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain COL) OX=93062 GN=sle1 PE=3 SV=1 |
| Q2G0U9 | 1.32e-13 | 34 | 221 | 19 | 202 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=sle1 PE=1 SV=1 |
| Q2FJH7 | 1.32e-13 | 34 | 221 | 19 | 202 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus (strain USA300) OX=367830 GN=sle1 PE=3 SV=1 |
| P0C1U7 | 1.32e-13 | 34 | 221 | 19 | 202 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus aureus OX=1280 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
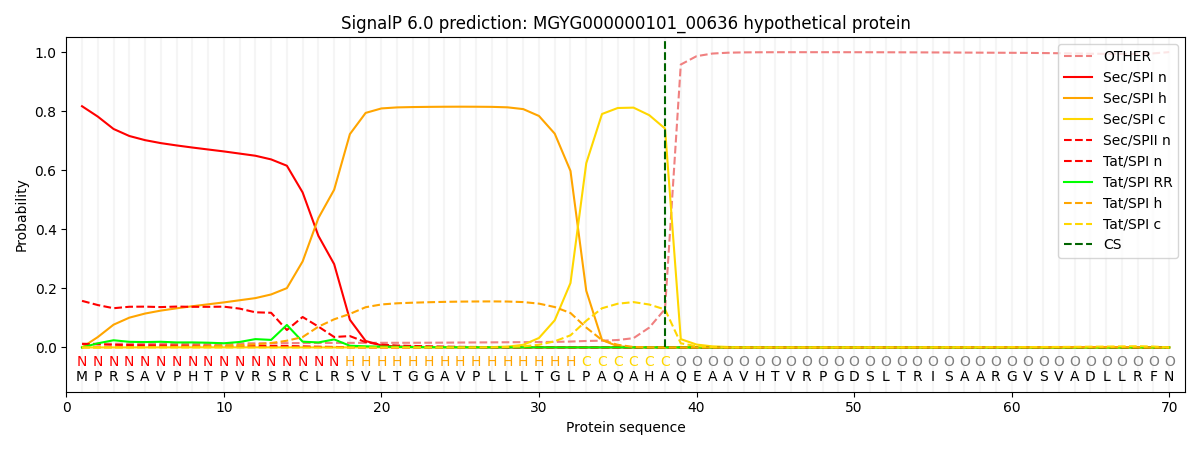
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.018914 | 0.802668 | 0.014517 | 0.161602 | 0.001861 | 0.000415 |
