You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000101_01886
You are here: Home > Sequence: MGYG000000101_01886
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
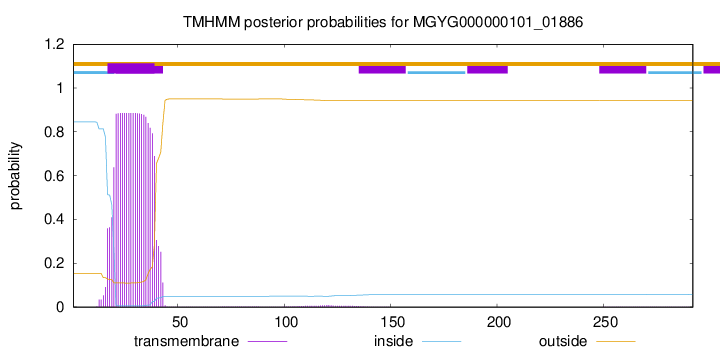
TMHMM annotations
Basic Information help
| Species | Micrococcus luteus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Micrococcaceae; Micrococcus; Micrococcus luteus | |||||||||||
| CAZyme ID | MGYG000000101_01886 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12024; End: 12902 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 1.48e-30 | 187 | 267 | 3 | 81 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK13914 | PRK13914 | 7.42e-21 | 188 | 275 | 381 | 463 | invasion associated endopeptidase. |
| COG0791 | Spr | 1.91e-19 | 175 | 277 | 77 | 181 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| NF033742 | NlpC_p60_RipB | 1.26e-17 | 188 | 266 | 92 | 179 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
| NF033741 | NlpC_p60_RipA | 8.61e-14 | 188 | 292 | 343 | 454 | NlpC/P60 family peptidoglycan endopeptidase RipA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGY89853.1 | 1.19e-181 | 1 | 292 | 1 | 292 |
| QGS21667.1 | 1.32e-178 | 1 | 292 | 1 | 292 |
| QGY83236.1 | 5.79e-178 | 1 | 292 | 1 | 294 |
| QDW16963.1 | 5.79e-178 | 1 | 292 | 1 | 294 |
| AWD24492.1 | 2.55e-176 | 1 | 292 | 1 | 292 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7CFL_A | 1.01e-13 | 172 | 277 | 16 | 120 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 4HPE_A | 5.78e-12 | 169 | 266 | 183 | 282 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 6B8C_A | 8.49e-12 | 177 | 266 | 32 | 119 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 2HBW_A | 5.38e-10 | 173 | 272 | 101 | 196 | ChainA, NLP/P60 protein [Trichormus variabilis ATCC 29413] |
| 2EVR_A | 6.02e-10 | 185 | 292 | 123 | 227 | ChainA, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102],2FG0_A Chain A, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102],2FG0_B Chain B, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31852 | 4.01e-20 | 49 | 292 | 159 | 414 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
| O07532 | 4.16e-18 | 45 | 277 | 238 | 471 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
| Q01839 | 4.08e-16 | 108 | 275 | 314 | 506 | Probable endopeptidase p60 OS=Listeria welshimeri OX=1643 GN=iap PE=3 SV=1 |
| P21171 | 5.14e-16 | 177 | 275 | 373 | 466 | Probable endopeptidase p60 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=iap PE=1 SV=2 |
| Q01836 | 9.01e-16 | 177 | 275 | 356 | 449 | Probable endopeptidase p60 OS=Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) OX=272626 GN=iap PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
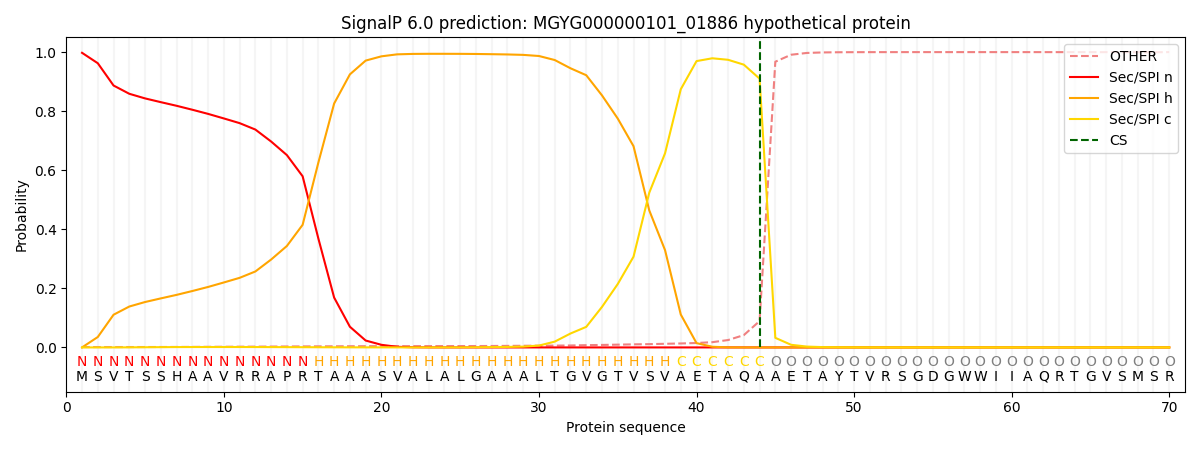
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002547 | 0.995002 | 0.000485 | 0.001436 | 0.000278 | 0.000237 |