You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000103_02406
You are here: Home > Sequence: MGYG000000103_02406
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
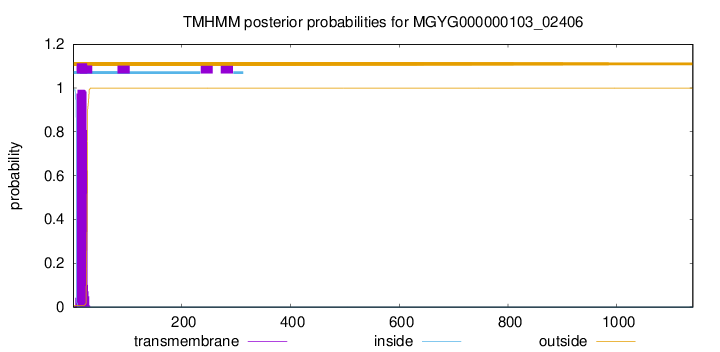
TMHMM annotations
Basic Information help
| Species | Bacilliculturomica massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Anaerovoracaceae; Bacilliculturomica; Bacilliculturomica massiliensis | |||||||||||
| CAZyme ID | MGYG000000103_02406 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 36556; End: 39978 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM54 | 206 | 317 | 2.7e-29 | 0.9649122807017544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033190 | inl_like_NEAT_1 | 2.32e-09 | 20 | 213 | 559 | 754 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 4.50e-06 | 162 | 203 | 1 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 5.89e-05 | 40 | 83 | 1 | 42 | S-layer homology domain. |
| PRK15319 | PRK15319 | 2.37e-04 | 532 | 888 | 119 | 437 | fibronectin-binding autotransporter adhesin ShdA. |
| PRK10811 | rne | 0.003 | 335 | 493 | 865 | 1019 | ribonuclease E; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIB69104.1 | 1.53e-117 | 5 | 392 | 4 | 391 |
| QHI71309.1 | 6.07e-110 | 5 | 427 | 4 | 407 |
| QOX65251.1 | 6.46e-82 | 1 | 386 | 3 | 390 |
| QRN84790.1 | 3.98e-78 | 8 | 371 | 9 | 367 |
| QIB70419.1 | 1.39e-76 | 1 | 427 | 1 | 432 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GZT_A | 2.34e-10 | 204 | 278 | 30 | 108 | CrystalStructure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38537 | 3.95e-14 | 45 | 243 | 40 | 239 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| C6CRV0 | 1.89e-10 | 13 | 207 | 1259 | 1455 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
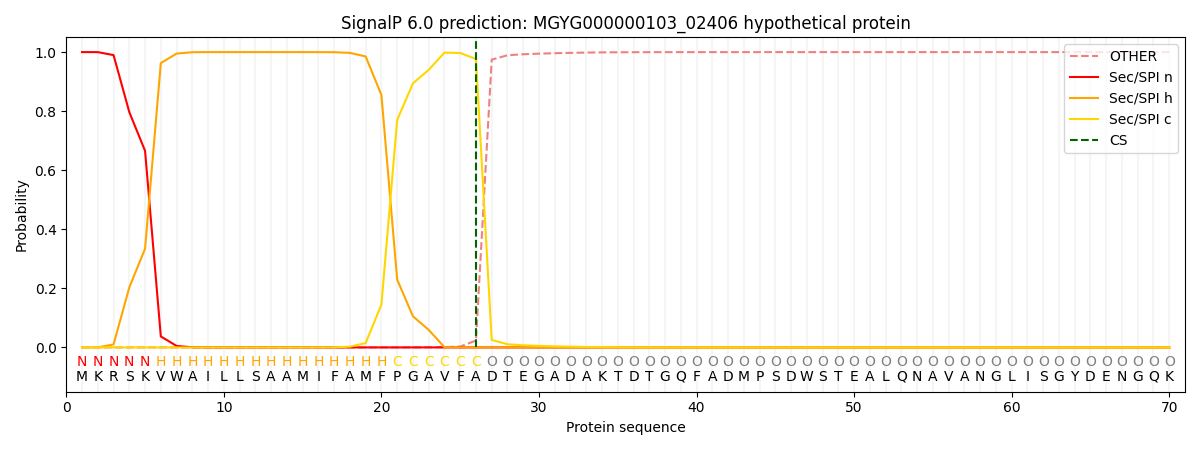
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000215 | 0.999146 | 0.000168 | 0.000169 | 0.000154 | 0.000141 |