You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000104_03076
You are here: Home > Sequence: MGYG000000104_03076
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
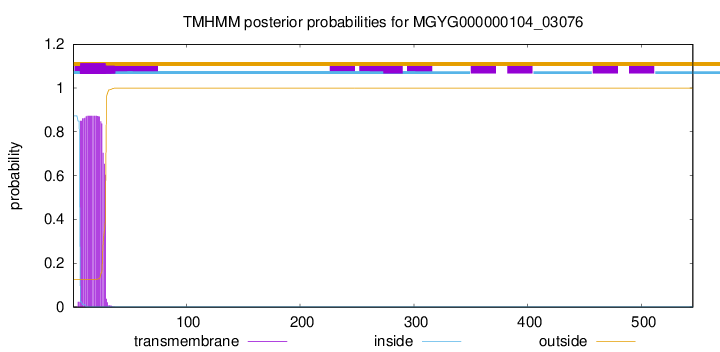
TMHMM annotations
Basic Information help
| Species | Clostridium cuniculi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium cuniculi | |||||||||||
| CAZyme ID | MGYG000000104_03076 | |||||||||||
| CAZy Family | CBM74 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24863; End: 26500 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM74 | 54 | 355 | 1.1e-108 | 0.9967948717948718 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK14081 | PRK14081 | 8.44e-22 | 365 | 535 | 288 | 463 | triple tyrosine motif-containing protein; Provisional |
| PRK14081 | PRK14081 | 6.58e-18 | 374 | 535 | 9 | 173 | triple tyrosine motif-containing protein; Provisional |
| PRK14081 | PRK14081 | 1.63e-06 | 397 | 534 | 514 | 653 | triple tyrosine motif-containing protein; Provisional |
| smart00089 | PKD | 4.88e-04 | 368 | 450 | 1 | 79 | Repeats in polycystic kidney disease 1 (PKD1) and other proteins. Polycystic kidney disease 1 protein contains 14 repeats, present elsewhere such as in microbial collagenases. |
| PRK14081 | PRK14081 | 0.002 | 336 | 434 | 360 | 457 | triple tyrosine motif-containing protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDM67953.1 | 4.71e-170 | 52 | 534 | 776 | 1261 |
| AKG25402.1 | 9.31e-101 | 54 | 358 | 1116 | 1416 |
| QIK84188.1 | 1.73e-91 | 54 | 358 | 853 | 1153 |
| QCT06902.1 | 2.95e-86 | 52 | 457 | 238 | 671 |
| AWX97480.1 | 6.99e-76 | 54 | 454 | 763 | 1163 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
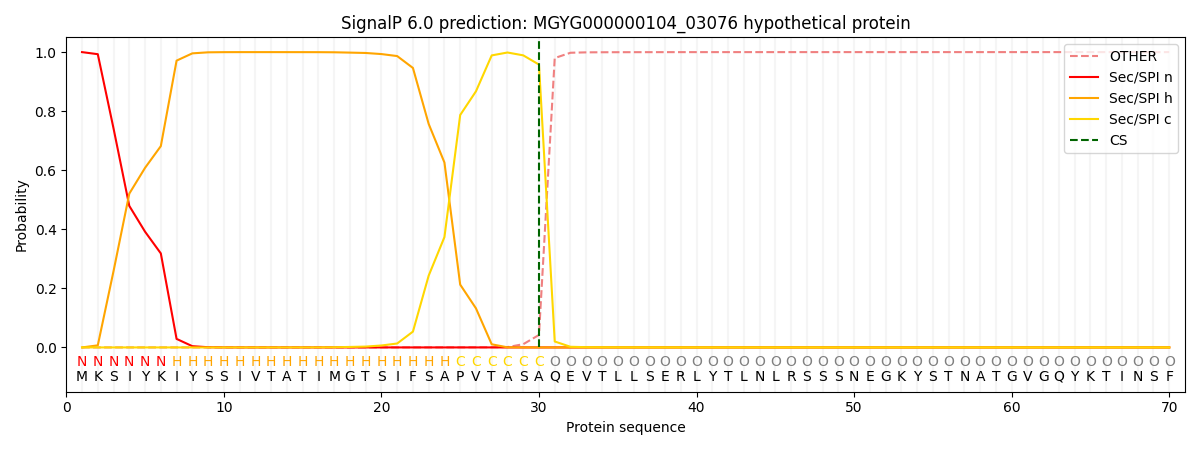
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000636 | 0.998497 | 0.000214 | 0.000249 | 0.000197 | 0.000182 |