You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000105_02263
You are here: Home > Sequence: MGYG000000105_02263
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides clarus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides clarus | |||||||||||
| CAZyme ID | MGYG000000105_02263 | |||||||||||
| CAZy Family | CE19 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 260844; End: 262007 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE19 | 57 | 386 | 2.1e-26 | 0.858433734939759 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0412 | DLH | 7.54e-20 | 110 | 345 | 13 | 186 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam12715 | Abhydrolase_7 | 6.51e-15 | 26 | 342 | 21 | 327 | Abhydrolase family. This is a family of probable bacterial abhydrolases. |
| COG1506 | DAP2 | 7.34e-11 | 91 | 345 | 358 | 579 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam01738 | DLH | 9.59e-07 | 112 | 345 | 1 | 170 | Dienelactone hydrolase family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37157.1 | 1.40e-182 | 18 | 382 | 364 | 727 |
| QEG26086.1 | 1.08e-22 | 62 | 381 | 26 | 327 |
| VTS00541.1 | 1.08e-22 | 62 | 381 | 26 | 327 |
| AWM40652.1 | 1.08e-22 | 62 | 381 | 26 | 327 |
| AMV30232.1 | 9.43e-19 | 62 | 384 | 25 | 329 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3G8Y_A | 2.67e-11 | 37 | 342 | 27 | 328 | ChainA, SusD/RagB-associated esterase-like protein [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34973 | 9.87e-25 | 62 | 381 | 2 | 297 | Putative hydrolase YtaP OS=Bacillus subtilis (strain 168) OX=224308 GN=ytaP PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
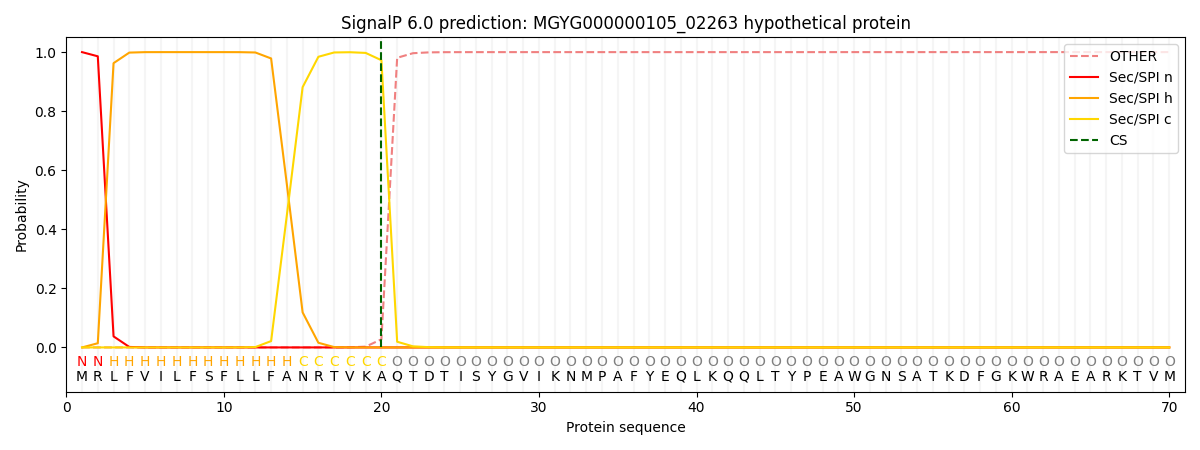
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000264 | 0.999045 | 0.000191 | 0.000150 | 0.000154 | 0.000140 |
