You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000105_02366
You are here: Home > Sequence: MGYG000000105_02366
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides clarus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides clarus | |||||||||||
| CAZyme ID | MGYG000000105_02366 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 382945; End: 384660 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 4 | 481 | 3.2e-79 | 0.8111111111111111 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 3.89e-32 | 5 | 529 | 7 | 529 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 1.73e-14 | 1 | 399 | 2 | 380 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 6.86e-05 | 62 | 180 | 2 | 119 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAJ74247.1 | 3.57e-29 | 24 | 371 | 28 | 357 |
| BBD06923.1 | 1.65e-25 | 10 | 396 | 19 | 396 |
| ACN14933.1 | 6.35e-25 | 2 | 363 | 7 | 347 |
| BBB93297.1 | 2.28e-24 | 11 | 393 | 12 | 364 |
| ACL74162.1 | 1.45e-23 | 1 | 388 | 14 | 369 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 5.22e-15 | 9 | 396 | 33 | 406 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O67270 | 1.97e-20 | 11 | 392 | 7 | 363 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| A8FRR0 | 3.22e-19 | 16 | 396 | 18 | 378 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1 |
| B5XTL1 | 2.40e-18 | 11 | 396 | 11 | 375 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Klebsiella pneumoniae (strain 342) OX=507522 GN=arnT PE=3 SV=1 |
| A6TF96 | 1.32e-17 | 11 | 396 | 11 | 375 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=arnT PE=3 SV=1 |
| C5BDQ8 | 2.33e-17 | 4 | 371 | 2 | 349 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999044 | 0.000905 | 0.000033 | 0.000005 | 0.000004 | 0.000022 |
TMHMM Annotations download full data without filtering help
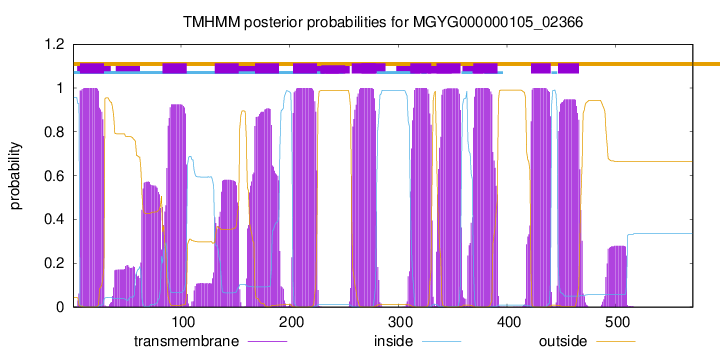
| start | end |
|---|---|
| 7 | 29 |
| 83 | 105 |
| 131 | 153 |
| 168 | 190 |
| 203 | 225 |
| 257 | 279 |
| 311 | 330 |
| 335 | 357 |
| 369 | 391 |
| 422 | 440 |
| 447 | 466 |
