You are browsing environment: HUMAN GUT
MGYG000000105_03273
Basic Information
help
Species
Bacteroides clarus
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides clarus
CAZyme ID
MGYG000000105_03273
CAZy Family
GH5
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000105
3966085
Isolate
Canada
North America
Gene Location
Start: 43405;
End: 45090
Strand: -
No EC number prediction in MGYG000000105_03273.
Family
Start
End
Evalue
family coverage
GH5
143
409
1.7e-23
0.7272727272727273
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam16586
DUF5060
8.78e-07
33
105
3
68
Domain of unknown function (DUF5060). This is the N-terminal domain of a putative glycoside hydrolase, DUF4038. It is found in a number of different bacterial orders.
more
COG3934
COG3934
4.22e-04
185
338
32
175
Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
Q5B833
2.75e-07
178
396
66
314
Mannan endo-1,4-beta-mannosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manB PE=1 SV=2
more
Q5AZ53
1.16e-06
267
396
159
315
Mannan endo-1,4-beta-mannosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manC PE=1 SV=1
more
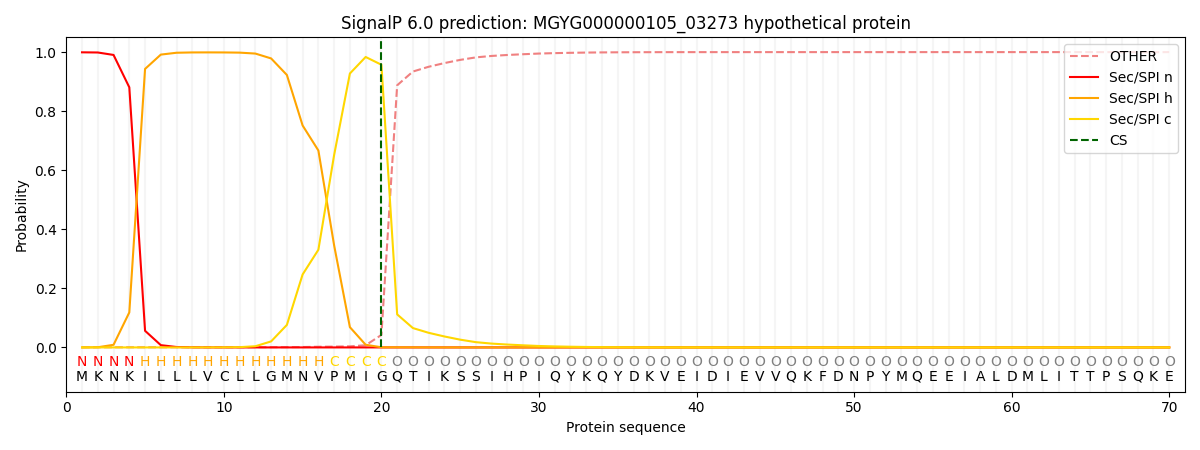
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.001748
0.997060
0.000551
0.000207
0.000201
0.000199
There is no transmembrane helices in MGYG000000105_03273.