You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000106_02235
You are here: Home > Sequence: MGYG000000106_02235
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
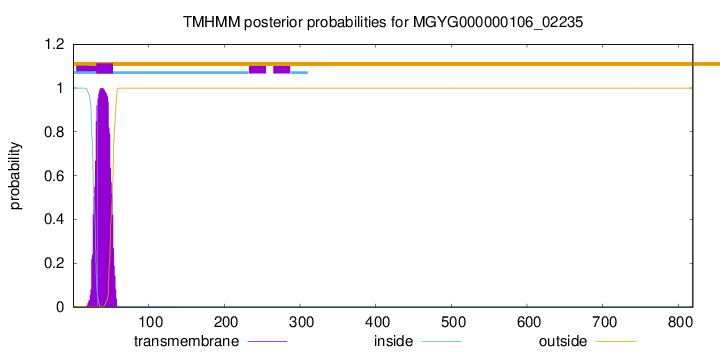
TMHMM annotations
Basic Information help
| Species | Enterococcus_D gallinarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Enterococcaceae; Enterococcus_D; Enterococcus_D gallinarum | |||||||||||
| CAZyme ID | MGYG000000106_02235 | |||||||||||
| CAZy Family | GT51 | |||||||||||
| CAZyme Description | Penicillin-binding protein 1A/1B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 356245; End: 358704 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT51 | 86 | 273 | 2.9e-53 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0744 | MrcB | 7.35e-156 | 23 | 740 | 1 | 654 | Membrane carboxypeptidase (penicillin-binding protein) [Cell wall/membrane/envelope biogenesis]. |
| COG5009 | MrcA | 3.56e-90 | 31 | 717 | 7 | 746 | Membrane carboxypeptidase/penicillin-binding protein [Cell wall/membrane/envelope biogenesis]. |
| pfam00912 | Transgly | 2.92e-61 | 86 | 274 | 1 | 177 | Transglycosylase. The penicillin-binding proteins are bifunctional proteins consisting of transglycosylase and transpeptidase in the N- and C-terminus respectively. The transglycosylase domain catalyzes the polymerization of murein glycan chains. |
| COG4953 | PbpC | 2.86e-47 | 54 | 724 | 16 | 591 | Membrane carboxypeptidase/penicillin-binding protein PbpC [Cell wall/membrane/envelope biogenesis]. |
| PRK11636 | mrcA | 9.93e-42 | 81 | 663 | 49 | 747 | penicillin-binding protein 1a; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOG27803.1 | 0.0 | 1 | 775 | 1 | 775 |
| AYY11009.1 | 0.0 | 1 | 775 | 1 | 775 |
| QGR81606.1 | 0.0 | 1 | 775 | 1 | 775 |
| QCT93040.1 | 0.0 | 1 | 775 | 1 | 775 |
| ATF70746.1 | 0.0 | 1 | 773 | 1 | 773 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JE5_A | 1.02e-218 | 48 | 770 | 3 | 718 | StructuralAnd Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6],2JE5_B Structural And Mechanistic Basis Of Penicillin Binding Protein Inhibition By Lactivicins [Streptococcus pneumoniae R6] |
| 2BG1_A | 7.96e-121 | 292 | 770 | 24 | 492 | Activesite restructuring regulates ligand recognition in classA Penicillin-binding proteins (PBPs) [Streptococcus pneumoniae R6],2XD5_A Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b [Streptococcus pneumoniae R6],2XD5_B Structural insights into the catalytic mechanism and the role of Streptococcus pneumoniae PBP1b [Streptococcus pneumoniae R6] |
| 2XD1_A | 7.96e-121 | 292 | 770 | 24 | 492 | ACTIVESITE RESTRUCTURING REGULATES LIGAND RECOGNITION IN CLASS A PENICILLIN-BINDING PROTEINS [Streptococcus pneumoniae R6],2XD1_B ACTIVE SITE RESTRUCTURING REGULATES LIGAND RECOGNITION IN CLASS A PENICILLIN-BINDING PROTEINS [Streptococcus pneumoniae R6] |
| 2UWX_A | 4.38e-120 | 292 | 770 | 24 | 492 | Activesite restructuring regulates ligand recognition in class A penicillin-binding proteins [Streptococcus pneumoniae R6] |
| 2Y2G_A | 2.41e-119 | 292 | 770 | 24 | 492 | Penicillin-BindingProtein 1b (Pbp-1b) In Complex With An Alkyl Boronate (A01) [Streptococcus pneumoniae R6],2Y2G_B Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (A01) [Streptococcus pneumoniae R6],2Y2H_A Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (Za2) [Streptococcus pneumoniae R6],2Y2H_B Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (Za2) [Streptococcus pneumoniae R6],2Y2I_A Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (Za3) [Streptococcus pneumoniae R6],2Y2J_A Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (Za4) [Streptococcus pneumoniae R6],2Y2K_A Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (Za5) [Streptococcus pneumoniae R6],2Y2L_A Penicillin-binding Protein 1b (pbp-1b) In Complex With An Alkyl Boronate (e06) [Streptococcus pneumoniae R6],2Y2L_B Penicillin-binding Protein 1b (pbp-1b) In Complex With An Alkyl Boronate (e06) [Streptococcus pneumoniae R6],2Y2M_A Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (E08) [Streptococcus pneumoniae R6],2Y2N_A Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (E07) [Streptococcus pneumoniae R6],2Y2O_A Penicillin-binding Protein 1b (pbp-1b) In Complex With An Alkyl Boronate (eo9) [Streptococcus pneumoniae R6],2Y2P_A Penicillin-binding protein 1b (pbp-1b) in complex with an alkyl boronate (z10) [Streptococcus pneumoniae R6],2Y2Q_A Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (Z06) [Streptococcus pneumoniae R6],2Y2Q_B Penicillin-Binding Protein 1b (Pbp-1b) In Complex With An Alkyl Boronate (Z06) [Streptococcus pneumoniae R6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0SRL7 | 3.25e-53 | 43 | 724 | 34 | 700 | Penicillin-binding protein 1A OS=Clostridium perfringens (strain SM101 / Type A) OX=289380 GN=pbpA PE=3 SV=1 |
| Q891X1 | 3.29e-53 | 84 | 725 | 64 | 694 | Penicillin-binding protein 1A OS=Clostridium tetani (strain Massachusetts / E88) OX=212717 GN=pbpA PE=3 SV=1 |
| A7FY32 | 3.97e-52 | 5 | 717 | 4 | 671 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain ATCC 19397 / Type A) OX=441770 GN=pbpA PE=3 SV=1 |
| A5I6G4 | 3.97e-52 | 5 | 717 | 4 | 671 | Penicillin-binding protein 1A OS=Clostridium botulinum (strain Hall / ATCC 3502 / NCTC 13319 / Type A) OX=441771 GN=pbpA PE=3 SV=1 |
| P39793 | 4.02e-52 | 9 | 713 | 22 | 665 | Penicillin-binding protein 1A/1B OS=Bacillus subtilis (strain 168) OX=224308 GN=ponA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
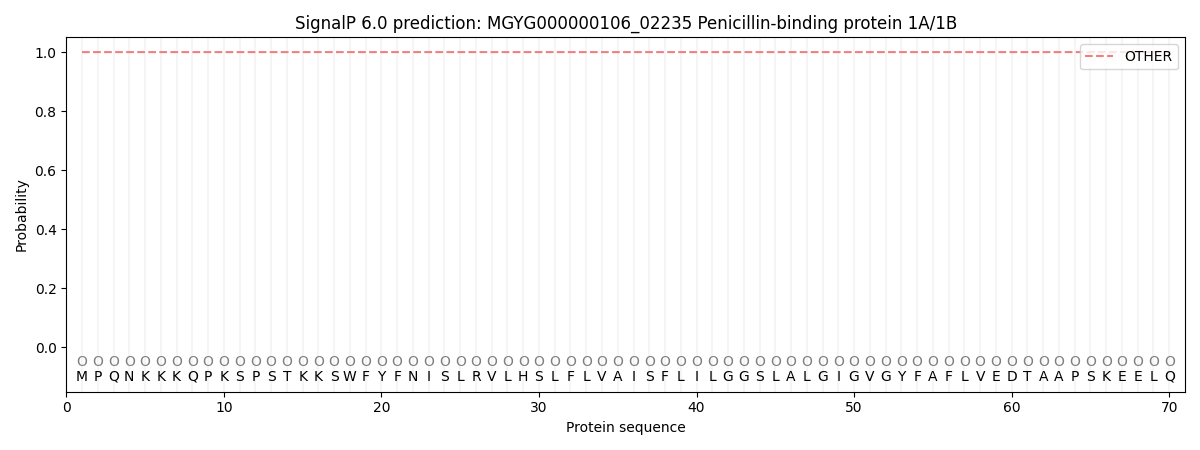
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000008 | 0.000007 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |