You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000111_01663
You are here: Home > Sequence: MGYG000000111_01663
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cutibacterium acnes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Cutibacterium; Cutibacterium acnes | |||||||||||
| CAZyme ID | MGYG000000111_01663 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 163553; End: 164932 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 75 | 398 | 1.7e-83 | 0.9869706840390879 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01301 | Glyco_hydro_35 | 7.85e-84 | 74 | 392 | 1 | 308 | Glycosyl hydrolases family 35. |
| PLN03059 | PLN03059 | 1.70e-31 | 68 | 384 | 30 | 323 | beta-galactosidase; Provisional |
| COG1874 | GanA | 3.77e-18 | 74 | 264 | 7 | 168 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 0.001 | 98 | 159 | 10 | 68 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam00150 | Cellulase | 0.002 | 95 | 277 | 21 | 198 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOH46078.1 | 1.71e-285 | 1 | 426 | 1 | 426 |
| BCY25970.1 | 9.83e-285 | 1 | 426 | 1 | 426 |
| ALU23968.1 | 2.28e-183 | 1 | 260 | 1 | 260 |
| QQO77160.1 | 4.51e-172 | 53 | 419 | 48 | 418 |
| ALT42701.1 | 8.93e-169 | 1 | 260 | 1 | 260 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1TG7_A | 2.27e-63 | 68 | 413 | 6 | 361 | Nativestructure of beta-galactosidase from Penicillium sp. [Penicillium sp.],1XC6_A Native Structure Of Beta-Galactosidase from Penicillium sp. in complex with Galactose [Penicillium sp.] |
| 5IFP_A | 2.66e-62 | 68 | 413 | 46 | 400 | StructureOf Beta-galactosidase From Aspergillus Niger [Aspergillus niger CBS 513.88] |
| 5IFT_A | 6.81e-62 | 68 | 413 | 46 | 400 | STRUCTUREOF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 3-b-Galactopyranosyl glucose [Aspergillus niger CBS 513.88],5IHR_A Structure Of E298q-beta-galactosidase From Aspergillus Niger In Complex With Allolactose [Aspergillus niger CBS 513.88],5JUV_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-b-Galactopyranosyl galactose [Aspergillus niger CBS 513.88],5MGC_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 4-Galactosyl-lactose [Aspergillus niger CBS 513.88],5MGD_A STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose [Aspergillus niger CBS 513.88] |
| 3OG2_A | 8.58e-59 | 68 | 423 | 26 | 387 | ChainA, Beta-galactosidase [Trichoderma reesei],3OGR_A Chain A, Beta-galactosidase [Trichoderma reesei],3OGS_A Chain A, Beta-galactosidase [Trichoderma reesei],3OGV_A Chain A, Beta-galactosidase [Trichoderma reesei] |
| 4IUG_A | 4.10e-58 | 68 | 411 | 46 | 399 | Crystalstructure of beta-galactosidase from Aspergillus oryzae in complex with galactose [Aspergillus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B0Y752 | 5.63e-68 | 61 | 403 | 25 | 378 | Probable beta-galactosidase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=lacC PE=3 SV=1 |
| Q4WNE4 | 5.63e-68 | 61 | 403 | 25 | 378 | Probable beta-galactosidase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=lacC PE=3 SV=1 |
| A1DM65 | 7.22e-67 | 61 | 403 | 25 | 378 | Probable beta-galactosidase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=lacC PE=3 SV=1 |
| Q2UMD5 | 9.29e-66 | 51 | 403 | 15 | 378 | Probable beta-galactosidase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=lacC PE=3 SV=1 |
| B8N2I5 | 9.29e-66 | 51 | 403 | 15 | 378 | Probable beta-galactosidase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=lacC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
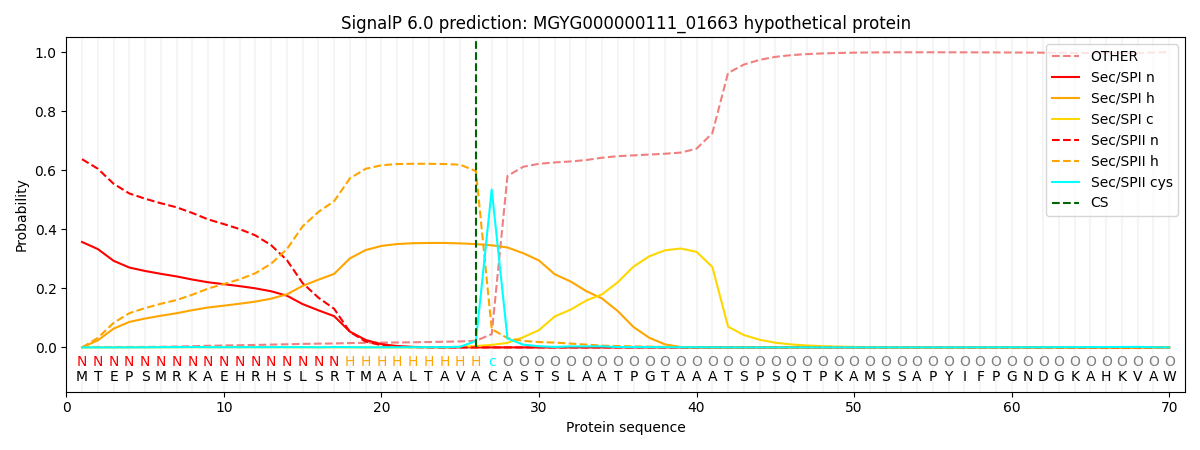
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002157 | 0.347366 | 0.645318 | 0.003125 | 0.001820 | 0.000204 |
