You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000111_02272
You are here: Home > Sequence: MGYG000000111_02272
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cutibacterium acnes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Propionibacteriaceae; Cutibacterium; Cutibacterium acnes | |||||||||||
| CAZyme ID | MGYG000000111_02272 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 155644; End: 157152 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 114 | 442 | 2.2e-81 | 0.9210526315789473 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 1.06e-87 | 107 | 449 | 1 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 2.94e-34 | 58 | 410 | 220 | 606 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam13088 | BNR_2 | 6.76e-14 | 131 | 436 | 1 | 280 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| cd18622 | GH32_Inu-like | 0.005 | 225 | 307 | 37 | 120 | glycoside hydrolase family 32 protein such as Aspergillus ficuum endo-inulinase (Inu2). This subfamily of glycosyl hydrolase family GH32 includes endo-inulinase (inu2, EC 3.2.1.7), exo-inulinase (Inu1, EC 3.2.1.80), invertase (EC 3.2.1.26), and levan fructotransferase (LftA, EC 4.2.2.16), among others. These enzymes cleave sucrose into fructose and glucose via beta-fructofuranosidase activity, producing invert sugar that is a mixture of dextrorotatory D-glucose and levorotatory D-fructose, thus named invertase (EC 3.2.1.26). These retaining enzymes (i.e. they retain the configuration at anomeric carbon atom of the substrate) catalyze hydrolysis in two steps involving a covalent glycosyl enzyme intermediate: an aspartate located close to the N-terminus acts as the catalytic nucleophile and a glutamate acts as the general acid/base; a conserved aspartate residue in the Arg-Asp-Pro (RDP) motif stabilizes the transition state. These enzymes are predicted to display a 5-fold beta-propeller fold as found for GH43 and CH68. The breakdown of sucrose is widely used as a carbon or energy source by bacteria, fungi, and plants. Invertase is used commercially in the confectionery industry, since fructose has a sweeter taste than sucrose and a lower tendency to crystallize. A common structural feature of all these enzymes is a 5-bladed beta-propeller domain, similar to GH43, that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALT35678.1 | 0.0 | 1 | 502 | 1 | 502 |
| AXM07790.1 | 0.0 | 1 | 502 | 1 | 502 |
| AER04689.1 | 0.0 | 1 | 502 | 1 | 502 |
| ALD70051.1 | 0.0 | 1 | 502 | 1 | 502 |
| ALU23799.1 | 0.0 | 1 | 502 | 1 | 502 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7LBU_A | 0.0 | 31 | 481 | 5 | 455 | ChainA, Exo-alpha-sialidase [Cutibacterium acnes],7LBV_A Chain A, Exo-alpha-sialidase [Cutibacterium acnes] |
| 1EUR_A | 8.79e-134 | 108 | 457 | 13 | 363 | Sialidase[Micromonospora viridifaciens],1EUS_A Sialidase Complexed With 2-Deoxy-2,3-Dehydro-N- Acetylneuraminic Acid [Micromonospora viridifaciens] |
| 1EUT_A | 1.89e-130 | 108 | 457 | 13 | 363 | Sialidase,Large 68kd Form, Complexed With Galactose [Micromonospora viridifaciens],1EUU_A Sialidase Or Neuraminidase, Large 68kd Form [Micromonospora viridifaciens] |
| 1WCQ_A | 6.70e-130 | 108 | 457 | 9 | 359 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BZD_A | 1.34e-129 | 108 | 457 | 9 | 359 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q02834 | 3.54e-129 | 108 | 457 | 55 | 405 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P29767 | 3.87e-19 | 105 | 444 | 371 | 814 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P62575 | 8.04e-16 | 228 | 440 | 538 | 769 | Sialidase A OS=Streptococcus pneumoniae OX=1313 GN=nanA PE=1 SV=1 |
| P62576 | 8.04e-16 | 228 | 440 | 538 | 769 | Sialidase A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=nanA PE=1 SV=1 |
| P15698 | 2.76e-12 | 119 | 448 | 53 | 390 | Sialidase OS=Paeniclostridium sordellii OX=1505 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
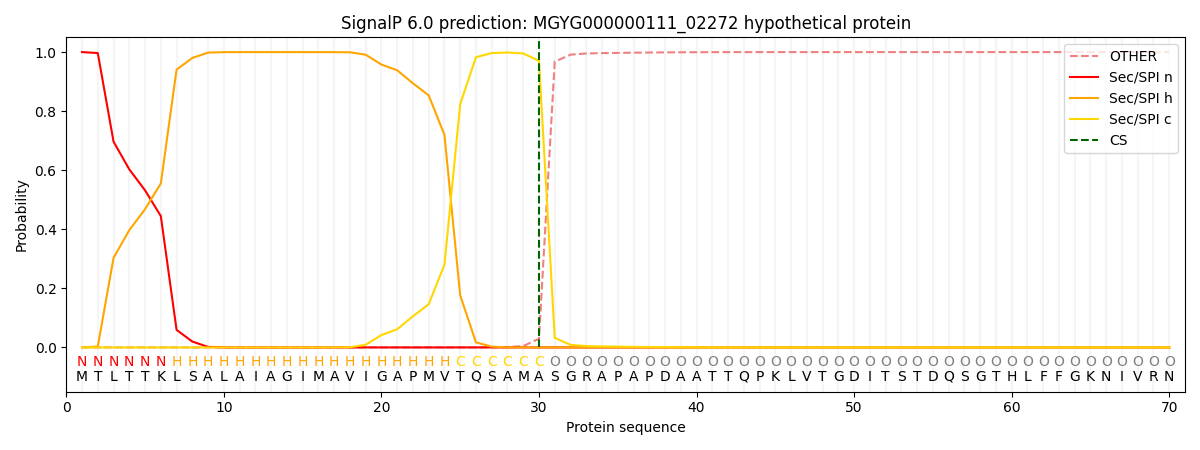
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000330 | 0.998949 | 0.000186 | 0.000214 | 0.000176 | 0.000149 |

