You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000117_00147
You are here: Home > Sequence: MGYG000000117_00147
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Barnesiella sp003150885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Barnesiellaceae; Barnesiella; Barnesiella sp003150885 | |||||||||||
| CAZyme ID | MGYG000000117_00147 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 143825; End: 145846 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 436 | 576 | 9.7e-18 | 0.7021276595744681 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08757 | CotH | 2.35e-38 | 76 | 416 | 1 | 318 | CotH kinase protein. Members of this family include the spore coat protein H (cotH). This protein is an atypical protein kinase that phosphorylates CotB and CotG. |
| pfam14200 | RicinB_lectin_2 | 1.24e-15 | 472 | 558 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 1.32e-11 | 437 | 570 | 2 | 126 | Ricin-type beta-trefoil lectin domain. |
| pfam14200 | RicinB_lectin_2 | 1.19e-09 | 516 | 576 | 1 | 58 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 8.89e-09 | 431 | 497 | 7 | 73 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI64517.1 | 2.30e-149 | 17 | 571 | 13 | 581 |
| QRR03748.1 | 2.46e-137 | 21 | 573 | 13 | 563 |
| APF19010.1 | 3.71e-49 | 7 | 471 | 5 | 485 |
| AEY66151.1 | 1.67e-47 | 35 | 512 | 44 | 508 |
| CUH93331.1 | 5.54e-40 | 33 | 422 | 250 | 701 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNC2 | 1.90e-06 | 438 | 672 | 294 | 516 | Endo-acting ulvan lyase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22190 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
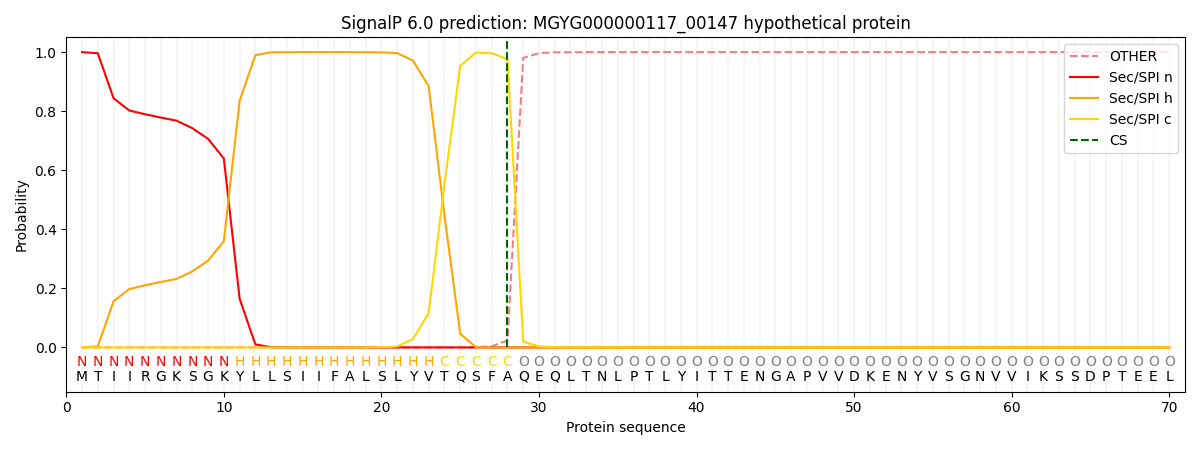
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000770 | 0.998374 | 0.000301 | 0.000179 | 0.000181 | 0.000173 |
