You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000117_00232
You are here: Home > Sequence: MGYG000000117_00232
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
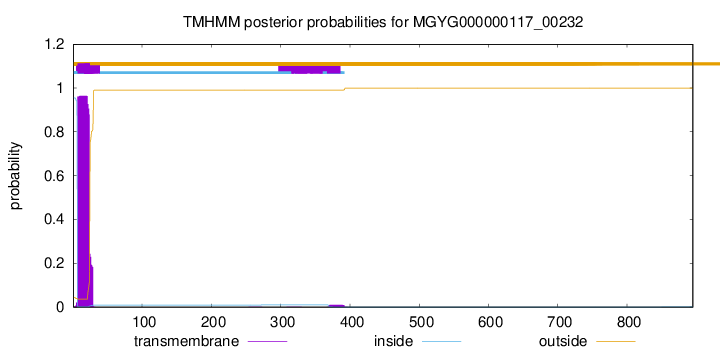
TMHMM annotations
Basic Information help
| Species | Barnesiella sp003150885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Barnesiellaceae; Barnesiella; Barnesiella sp003150885 | |||||||||||
| CAZyme ID | MGYG000000117_00232 | |||||||||||
| CAZy Family | CBM35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 255947; End: 258634 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM35 | 753 | 871 | 2.2e-18 | 0.9243697478991597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04083 | CBM35_Lmo2446-like | 4.77e-23 | 753 | 870 | 3 | 116 | Carbohydrate Binding Module 35 (CBM35) domains similar to Lmo2446. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes. Some CBM35 domains belonging to this family are appended to glycoside hydrolase (GH) family domains, including glycoside hydrolase family 31 (GH31), for example the CBM35 domain of Lmo2446, an uncharacterized protein from Listeria monocytogenes EGD-e. These CBM35s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. GH31 has a wide range of hydrolytic activities such as alpha-glucosidase, alpha-xylosidase, 6-alpha-glucosyltransferase, or alpha-1,4-glucan lyase, cleaving a terminal carbohydrate moiety from a substrate that may be a starch or a glycoprotein. Most characterized GH31 enzymes are alpha-glucosidases. |
| COG3408 | GDB1 | 1.71e-16 | 209 | 616 | 232 | 641 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| cd04082 | CBM35_pectate_lyase-like | 1.64e-13 | 752 | 863 | 2 | 109 | Carbohydrate Binding Module family 35 (CBM35), pectate lyase-like; appended mainly to enzymes that bind mannan (Man), xylan, glucuronic acid (GlcA) and possibly glucans. This family includes carbohydrate binding module family 35 (CBM35) domains that are non-catalytic carbohydrate binding domains that are appended mainly to enzymes that bind mannan (Man), xylan, glucuronic acid (GlcA) and possibly glucans. Included in this family are CBM35s of pectate lyases, including pectate lyase 10A from Cellvibrio japonicas, these enzymes release delta-4,5-anhydrogalaturonic acid (delta4,5-GalA) from pectin, thus identifying a signature molecule for plant cell wall degradation. CBM35s are unique in that they display conserved specificity through extensive sequence similarity but divergent function through their appended catalytic modules. They are known to bind alpha-D-galactose (Gal), mannan (Man), xylan, glucuronic acid (GlcA), a beta-polymer of mannose, and possibly glucans, forming four subfamilies based on general ligand specificities (galacto, urono, manno, and gluco configurations). In contrast to most CBMs that are generally rigid proteins, CBM35 undergoes significant conformational change upon ligand binding. Some CBM35s bind their ligands in a calcium-dependent manner, especially those binding uronic acids. |
| cd02795 | CBM6-CBM35-CBM36_like | 4.15e-09 | 752 | 870 | 1 | 116 | Carbohydrate Binding Module 6 (CBM6) and CBM35_like superfamily. Carbohydrate binding module family 6 (CBM6, family 6 CBM), also known as cellulose binding domain family VI (CBD VI), and related CBMs (CBM35 and CBM36). These are non-catalytic carbohydrate binding domains found in a range of enzymes that display activities against a diverse range of carbohydrate targets, including mannan, xylan, beta-glucans, cellulose, agarose, and arabinans. These domains facilitate the strong binding of the appended catalytic modules to their dedicated, insoluble substrates. Many of these CBMs are associated with glycoside hydrolase (GH) domains. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. CBM36s are calcium-dependent xylan binding domains. CBM35s display conserved specificity through extensive sequence similarity, but divergent function through their appended catalytic modules. This alignment model also contains the C-terminal domains of bacterial insecticidal toxins, where they may be involved in determining insect specificity through carbohydrate binding functionality. |
| cd04086 | CBM35_mannanase-like | 5.71e-09 | 751 | 863 | 1 | 105 | Carbohydrate Binding Module 35 (CBM35); appended to several carbohydrate binding enzymes, including several glycoside hydrolase (GH) family 26 mannanase domains. This family includes carbohydrate binding module 35 (CBM35) domains that are appended to several carbohydrate binding enzymes, including periplasmic component of ABC-type sugar transport system involved in carbohydrate transport and metabolism, and several glycoside hydrolase (GH) domains, including GH26. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB35s belonging to this family are mannanase A from Clostridium thermocellum (GH26), Man26B from Paenibacillus sp. BME-14 (GH26), and the multifunctional Cel44C-Man26A from Paenibacillus polymyxa GS01 (which has two GH domains, GH44 and GH26). GH26 mainly includes mannan endo-1,4-beta-mannosidase which hydrolyzes 1,4-beta-D-linkages in mannans, galacto-mannans, glucomannans, and galactoglucomannans, but displays little activity towards other plant cell wall polysaccharides. A few proteins belonging to this family have additional CBM3 domains; these CBM3s are not found in the CBM6-CBM35-CBM36_like superfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75824.1 | 0.0 | 16 | 895 | 12 | 887 |
| QEC45824.1 | 0.0 | 13 | 893 | 5 | 885 |
| AFG37033.1 | 3.58e-146 | 183 | 895 | 61 | 782 |
| QEC58493.1 | 2.78e-136 | 205 | 895 | 199 | 877 |
| QEN03989.1 | 1.65e-134 | 169 | 879 | 46 | 761 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5X7G_A | 1.07e-11 | 753 | 870 | 405 | 518 | CrystalStructure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase [Paenibacillus sp. 598K],5X7H_A Crystal Structure of Paenibacillus sp. 598K cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltoheptaose [Paenibacillus sp. 598K] |
| 3WNL_A | 2.17e-10 | 753 | 870 | 387 | 500 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNM_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNN_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_A Chain A, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNO_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNK_A | 2.19e-10 | 753 | 870 | 406 | 519 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 3WNP_A | 4.43e-09 | 753 | 870 | 387 | 500 | ChainA, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans],3WNP_B Chain B, Cycloisomaltooligosaccharide glucanotransferase [Niallia circulans] |
| 5X7O_A | 4.79e-08 | 739 | 867 | 822 | 951 | Crystalstructure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase [Paenibacillus sp. 598K],5X7O_B Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase [Paenibacillus sp. 598K],5X7P_A Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with acarbose [Paenibacillus sp. 598K],5X7P_B Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with acarbose [Paenibacillus sp. 598K],5X7Q_A Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose [Paenibacillus sp. 598K],5X7Q_B Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose [Paenibacillus sp. 598K],5X7R_A Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with isomaltohexaose [Paenibacillus sp. 598K],5X7R_B Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with isomaltohexaose [Paenibacillus sp. 598K],5X7S_A Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative [Paenibacillus sp. 598K],5X7S_B Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative [Paenibacillus sp. 598K] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P70873 | 3.41e-12 | 753 | 870 | 415 | 528 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 GN=cit PE=3 SV=1 |
| P94286 | 1.38e-09 | 753 | 870 | 423 | 536 | Cycloisomaltooligosaccharide glucanotransferase OS=Niallia circulans OX=1397 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
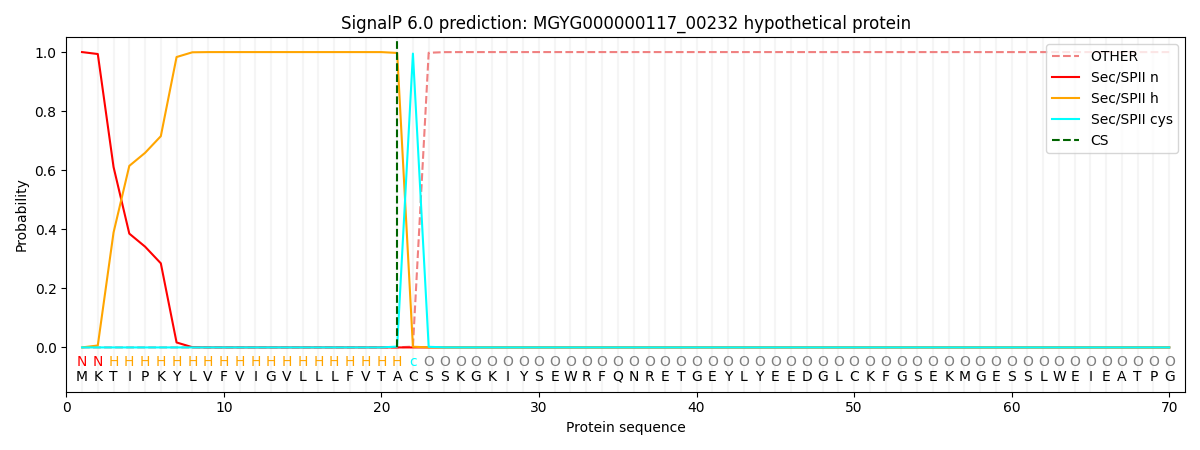
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000073 | 0.000000 | 0.000000 | 0.000000 |