You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000122_00091
You are here: Home > Sequence: MGYG000000122_00091
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
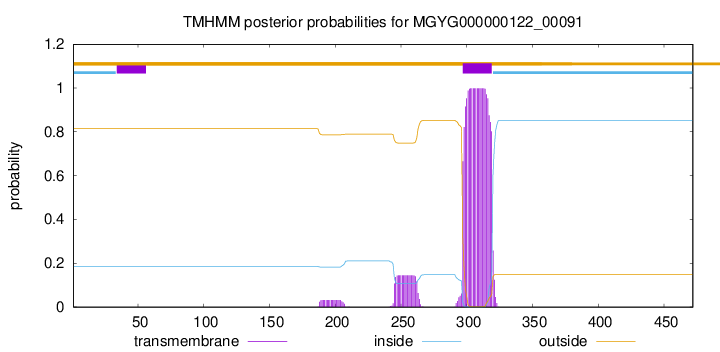
TMHMM annotations
Basic Information help
| Species | Staphylococcus epidermidis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus epidermidis | |||||||||||
| CAZyme ID | MGYG000000122_00091 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 120295; End: 121713 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033598 | elast_bind_EbpS | 2.78e-52 | 15 | 471 | 1 | 466 | elastin-binding protein EbpS. The elastin-binding protein EbpS is an adhesin described in Staphylococcus aureus, with orthologs found in many additional staphylococcal species. EbpS is a membrane protein that lacks an N-terminal signal peptide region, has extensive regions low-complexity sequence rich in Asn and Gln, and has a C-terminal LysM domain. |
| pfam01476 | LysM | 1.04e-05 | 430 | 471 | 7 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARG66600.1 | 3.41e-146 | 1 | 472 | 1 | 472 |
| AVG09544.1 | 3.41e-146 | 1 | 472 | 1 | 472 |
| AVA12093.1 | 1.95e-145 | 1 | 472 | 1 | 472 |
| VEJ61278.1 | 1.95e-145 | 1 | 472 | 1 | 472 |
| AUJ73827.1 | 1.95e-145 | 1 | 472 | 1 | 472 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8CP59 | 5.34e-138 | 13 | 472 | 1 | 460 | Probable elastin-binding protein EbpS OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=ebpS PE=4 SV=2 |
| Q5HP65 | 3.04e-137 | 13 | 472 | 1 | 460 | Probable elastin-binding protein EbpS OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=ebpS PE=4 SV=1 |
| Q53630 | 3.94e-25 | 285 | 471 | 308 | 486 | Elastin-binding protein EbpS OS=Staphylococcus aureus OX=1280 GN=ebpS PE=1 SV=4 |
| Q6GGT1 | 3.94e-25 | 285 | 471 | 308 | 486 | Elastin-binding protein EbpS OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=ebpS PE=3 SV=3 |
| Q2YY76 | 1.73e-24 | 285 | 471 | 305 | 483 | Elastin-binding protein EbpS OS=Staphylococcus aureus (strain bovine RF122 / ET3-1) OX=273036 GN=ebpS PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
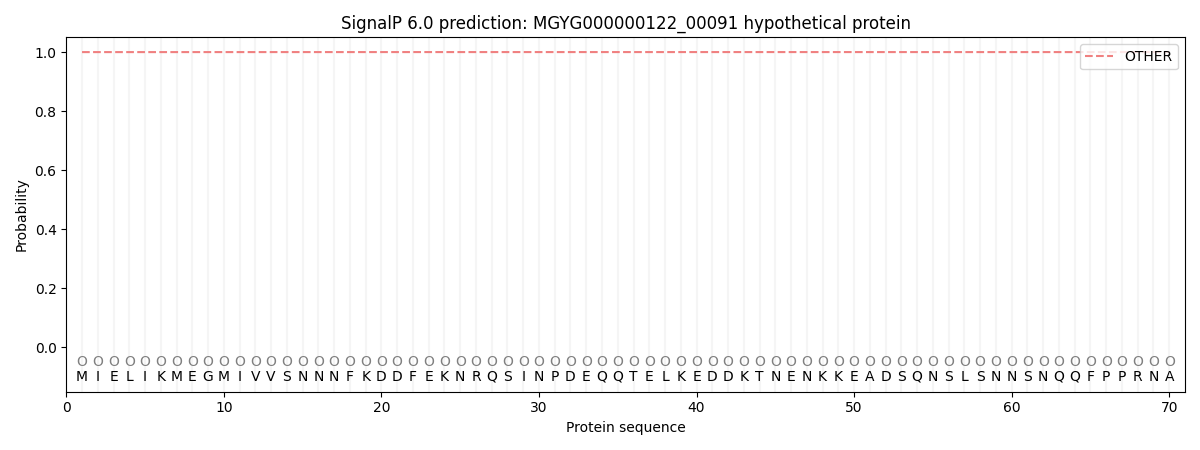
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000057 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |