You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000122_02363
You are here: Home > Sequence: MGYG000000122_02363
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus epidermidis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus epidermidis | |||||||||||
| CAZyme ID | MGYG000000122_02363 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12335; End: 17095 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR01760 | tape_meas_TP901 | 4.16e-77 | 165 | 510 | 1 | 348 | phage tail tape measure protein, TP901 family, core region. This model represents a reasonably well conserved core region of a family of phage tail proteins. The member from phage TP901-1 was characterized as a tail length tape measure protein in that a shortened form of the protein leads to phage with proportionately shorter tails. [Mobile and extrachromosomal element functions, Prophage functions] |
| pfam10145 | PhageMin_Tail | 1.95e-61 | 206 | 403 | 3 | 200 | Phage-related minor tail protein. Members of this family are found in putative phage tail tape measure proteins. |
| COG3953 | SLT | 2.50e-44 | 1371 | 1553 | 17 | 196 | SLT domain protein [Mobilome: prophages, transposons]. |
| cd13402 | LT_TF-like | 1.20e-38 | 1372 | 1489 | 4 | 116 | lytic transglycosylase-like domain of tail fiber-like proteins and similar domains. These tail fiber-like proteins are multi-domain proteins that include a lytic transglycosylase (LT) domain. Members of the LT family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, and the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG5412 | COG5412 | 1.38e-26 | 499 | 1053 | 1 | 496 | Phage-related protein [Mobilome: prophages, transposons]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTN13679.1 | 9.54e-295 | 160 | 1556 | 163 | 1447 |
| QUM70332.1 | 1.50e-289 | 1 | 1550 | 1 | 1595 |
| QUM67886.1 | 4.52e-288 | 1 | 1550 | 1 | 1595 |
| QCT63197.1 | 1.58e-271 | 159 | 1556 | 162 | 1518 |
| ALF33686.1 | 1.58e-271 | 159 | 1556 | 162 | 1518 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5KQB_A | 2.78e-11 | 1228 | 1314 | 27 | 118 | Identificationand structural characterization of LytU [Staphylococcus aureus],5KQC_A Identification and structural characterization of LytU [Staphylococcus aureus] |
| 5NMY_A | 9.64e-08 | 1226 | 1343 | 27 | 145 | NMRsolution structure of lysostaphin [Staphylococcus simulans] |
| 4QP5_A | 1.02e-07 | 1226 | 1335 | 29 | 140 | Catalyticdomain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the presence of phosphate [Staphylococcus simulans bv. staphylolyticus],4QP5_B Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the presence of phosphate [Staphylococcus simulans bv. staphylolyticus],4QPB_A Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate [Staphylococcus simulans],4QPB_B Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate [Staphylococcus simulans] |
| 4LXC_A | 1.08e-07 | 1226 | 1343 | 29 | 147 | Theantimicrobial peptidase lysostaphin from Staphylococcus simulans [Staphylococcus simulans],4LXC_B The antimicrobial peptidase lysostaphin from Staphylococcus simulans [Staphylococcus simulans],4LXC_C The antimicrobial peptidase lysostaphin from Staphylococcus simulans [Staphylococcus simulans],4LXC_D The antimicrobial peptidase lysostaphin from Staphylococcus simulans [Staphylococcus simulans] |
| 6RK4_A | 3.29e-07 | 1226 | 1343 | 275 | 393 | LysostaphinSH3b P4-G5 complex, synchrotron dataset [Staphylococcus simulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E7DNB6 | 5.41e-66 | 158 | 512 | 50 | 404 | Tape measure protein OS=Pneumococcus phage Dp-1 OX=59241 GN=TMP PE=4 SV=1 |
| O64314 | 1.65e-26 | 181 | 504 | 214 | 542 | Probable tape measure protein OS=Escherichia phage P2 OX=10679 GN=T PE=3 SV=1 |
| Q24LI1 | 1.23e-15 | 166 | 654 | 62 | 563 | Probable tape measure protein OS=Clostridium phage phiCD119 (strain Clostridium difficile/United States/Govind/2006) OX=1283341 PE=3 SV=1 |
| P45931 | 4.45e-11 | 1372 | 1486 | 1367 | 1483 | Uncharacterized protein YqbO OS=Bacillus subtilis (strain 168) OX=224308 GN=yqbO PE=1 SV=2 |
| O05156 | 3.08e-07 | 1226 | 1337 | 146 | 259 | Glycyl-glycine endopeptidase ALE-1 OS=Staphylococcus capitis OX=29388 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
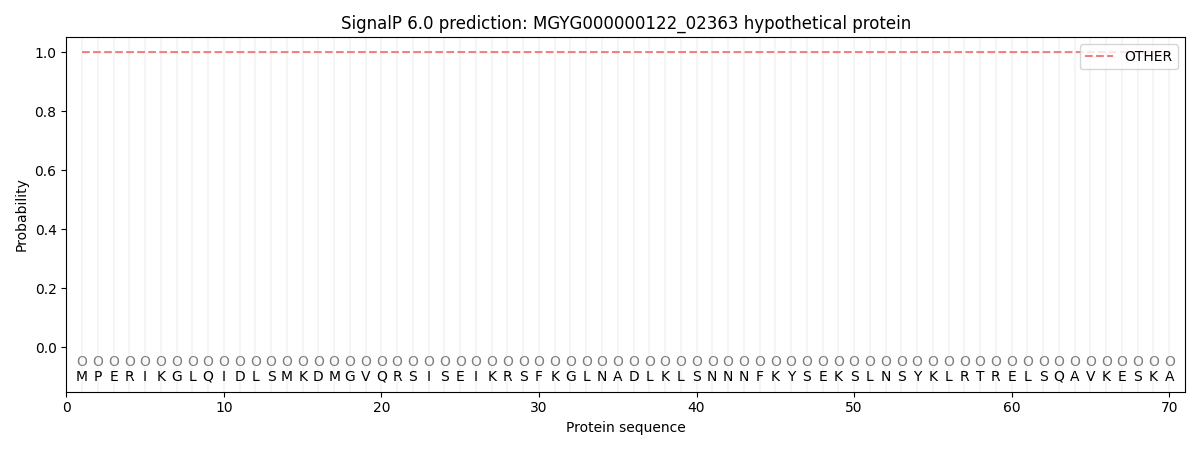
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000067 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
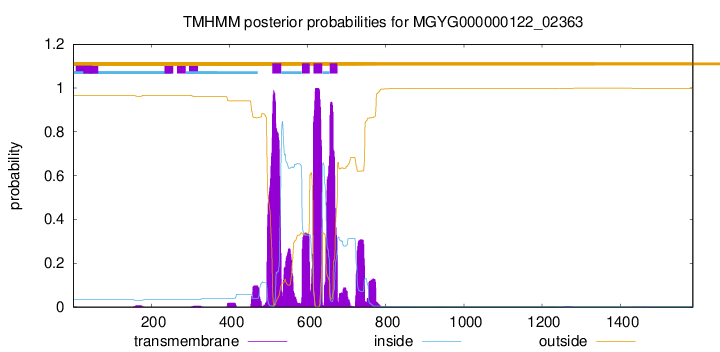
| start | end |
|---|---|
| 510 | 532 |
| 586 | 605 |
| 615 | 637 |
| 657 | 676 |
