You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000122_02384
You are here: Home > Sequence: MGYG000000122_02384
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
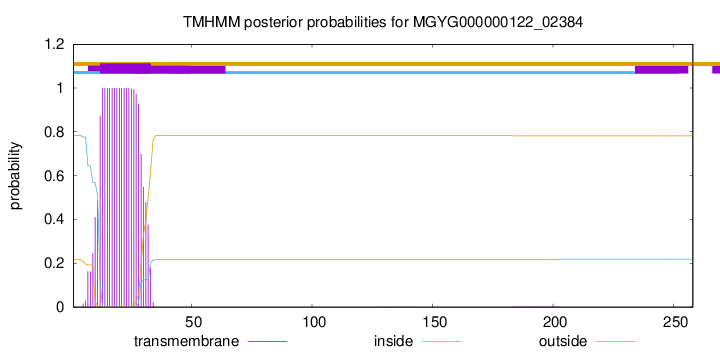
TMHMM annotations
Basic Information help
| Species | Staphylococcus epidermidis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus epidermidis | |||||||||||
| CAZyme ID | MGYG000000122_02384 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8243; End: 9019 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 115 | 241 | 8.1e-24 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.00e-92 | 15 | 258 | 2 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 9.30e-35 | 103 | 244 | 2 | 142 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam01832 | Glucosaminidase | 1.33e-10 | 114 | 201 | 1 | 78 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| COG1705 | FlgJ | 0.002 | 105 | 185 | 34 | 116 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| NF033232 | small_YtzI | 0.006 | 9 | 40 | 1 | 32 | YtzI protein. Members of this family include YtzI from Bacillus subtilis, and homologs widely distributed in the Firmicutes. The pattern of sequence conservation suggests the protein begins with a hydrophobic stretch without any basic residue near the initiator Met residue. Members of this family average about 53 residues in length. At the time this model was constructed, members were included in Pfam model PF12606.6 (Tumour necrosis factor receptor superfamily member 19), seemingly in error. This model was constructed to separate the two families. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGY92388.1 | 5.46e-186 | 1 | 258 | 1 | 258 |
| QRJ50720.1 | 5.46e-186 | 1 | 258 | 1 | 258 |
| VEJ60718.1 | 5.46e-186 | 1 | 258 | 1 | 258 |
| QRL26207.1 | 5.46e-186 | 1 | 258 | 1 | 258 |
| QRJ31569.1 | 5.46e-186 | 1 | 258 | 1 | 258 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PI7_A | 1.40e-111 | 35 | 258 | 5 | 228 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 1.15e-110 | 35 | 258 | 5 | 228 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 7.68e-52 | 61 | 246 | 35 | 232 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 1.89e-43 | 66 | 255 | 79 | 285 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 6FXP_A | 2.45e-43 | 66 | 242 | 49 | 230 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O33635 | 7.92e-49 | 29 | 246 | 1088 | 1323 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 7.92e-49 | 29 | 246 | 1088 | 1323 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q8CPQ1 | 7.92e-49 | 29 | 246 | 1088 | 1323 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| Q8NX96 | 4.30e-47 | 29 | 246 | 1009 | 1244 | Bifunctional autolysin OS=Staphylococcus aureus (strain MW2) OX=196620 GN=atl PE=3 SV=1 |
| Q6GAG0 | 4.30e-47 | 29 | 246 | 1003 | 1238 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
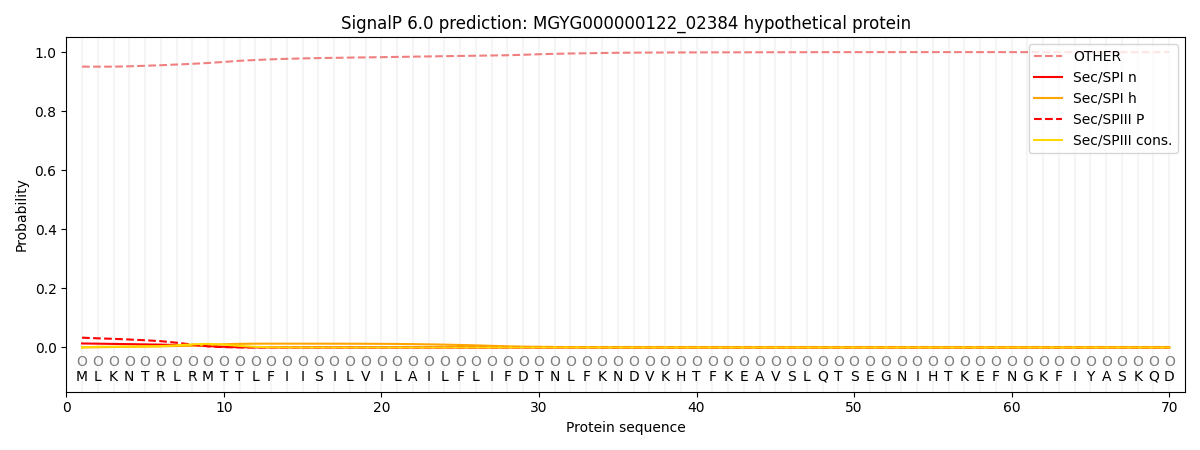
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.951373 | 0.012299 | 0.003006 | 0.000094 | 0.000052 | 0.033197 |