You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000123_02824
You are here: Home > Sequence: MGYG000000123_02824
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
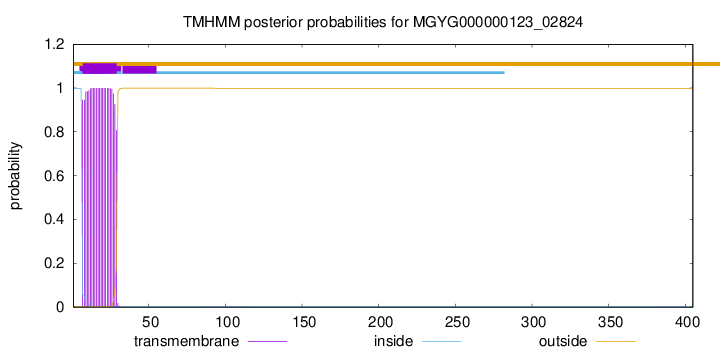
TMHMM annotations
Basic Information help
| Species | Blautia sp001304935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia; Blautia sp001304935 | |||||||||||
| CAZyme ID | MGYG000000123_02824 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 161419; End: 162636 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH25 | 39 | 213 | 2.7e-47 | 0.9943502824858758 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06525 | GH25_Lyc-like | 7.57e-72 | 37 | 222 | 1 | 184 | Lyc muramidase is an autolytic lysozyme (autolysin) from Clostridium acetobutylicum encoded by the lyc gene. Lyc has a glycosyl hydrolase family 25 (GH25) catalytic domain. Endo-N-acetylmuramidases are lysozymes (also referred to as peptidoglycan hydrolases) that degrade bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues. |
| cd00599 | GH25_muramidase | 1.65e-48 | 37 | 222 | 1 | 186 | Endo-N-acetylmuramidases (muramidases) are lysozymes (also referred to as peptidoglycan hydrolases) that degrade bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues. This family of muramidases contains a glycosyl hydrolase family 25 (GH25) catalytic domain and is found in bacteria, fungi, slime molds, round worms, protozoans and bacteriophages. The bacteriophage members are referred to as endolysins which are involved in lysing the host cell at the end of the replication cycle to allow release of mature phage particles. Endolysins are typically modular enzymes consisting of a catalytically active domain that hydrolyzes the peptidoglycan cell wall and a cell wall-binding domain that anchors the protein to the cell wall. Endolysins generally have narrow substrate specificities with either intra-species or intra-genus bacteriolytic activity. |
| pfam01183 | Glyco_hydro_25 | 5.38e-45 | 39 | 213 | 1 | 180 | Glycosyl hydrolases family 25. |
| cd06414 | GH25_LytC-like | 1.89e-42 | 36 | 220 | 1 | 188 | The LytC lysozyme of Streptococcus pneumoniae is a bacterial cell wall hydrolase that cleaves the beta1-4-glycosydic bond located between the N-acetylmuramoyl-N-glucosaminyl residues of the cell wall polysaccharide chains. LytC is composed of a C-terminal glycosyl hydrolase family 25 (GH25) domain and an N-terminal choline-binding module (CBM) consisting of eleven homologous repeats that specifically recognizes the choline residues of pneumococcal lipoteichoic and teichoic acids. This domain arrangement is the reverse of the major pneumococcal autolysin, LytA, and the CPL-1-like lytic enzymes of the pneumococcal bacteriophages, in which the CBM (consisting of six repeats) is at the C-terminus. This model represents the C-terminal catalytic domain of the LytC-like enzymes. |
| cd06524 | GH25_YegX-like | 1.05e-38 | 38 | 222 | 2 | 192 | YegX is an uncharacterized bacterial protein with a glycosyl hydrolase family 25 (GH25) catalytic domain that is similar in sequence to the CH-type (Chalaropsis-type) lysozymes of the GH25 family of endolysins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBE96304.1 | 4.00e-250 | 3 | 405 | 1 | 403 |
| QIB56830.1 | 8.07e-250 | 3 | 405 | 1 | 403 |
| QMW80393.1 | 8.07e-250 | 3 | 405 | 1 | 403 |
| ANU77129.1 | 9.52e-242 | 3 | 405 | 1 | 403 |
| QJU17248.1 | 9.52e-242 | 3 | 405 | 1 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4KRU_A | 3.78e-43 | 37 | 231 | 21 | 216 | X-raystructure of catalytic domain of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101] |
| 4KRT_A | 8.78e-42 | 37 | 231 | 21 | 216 | X-raystructure of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101],4KRT_B X-ray structure of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101] |
| 5A6S_A | 1.97e-24 | 28 | 224 | 14 | 203 | Crystalstructure of the CTP1L endolysin reveals how its activity is regulated by a secondary translation product [Clostridium phage phiCTP1] |
| 1JFX_A | 1.30e-18 | 32 | 222 | 1 | 202 | Crystalstructure of the bacterial lysozyme from Streptomyces coelicolor at 1.65 A resolution [Streptomyces coelicolor] |
| 4B8V_A | 3.80e-15 | 244 | 400 | 38 | 212 | ChainA, Extracellular Protein 6 [Fulvia fulva],4B9H_A Chain A, Extracellular Protein 6 [Fulvia fulva] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26836 | 1.93e-41 | 28 | 224 | 1 | 198 | Probable autolytic lysozyme OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=lyc PE=3 SV=2 |
| P34020 | 9.08e-28 | 37 | 228 | 2 | 186 | Autolytic lysozyme OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=lyc PE=1 SV=1 |
| P76421 | 7.14e-24 | 6 | 222 | 41 | 254 | Uncharacterized protein YegX OS=Escherichia coli (strain K12) OX=83333 GN=yegX PE=3 SV=2 |
| Q8FFY2 | 9.83e-24 | 6 | 222 | 41 | 254 | Uncharacterized protein YegX OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=yegX PE=3 SV=2 |
| Q8X7H0 | 9.83e-24 | 6 | 222 | 41 | 254 | Uncharacterized protein YegX OS=Escherichia coli O157:H7 OX=83334 GN=yegX PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
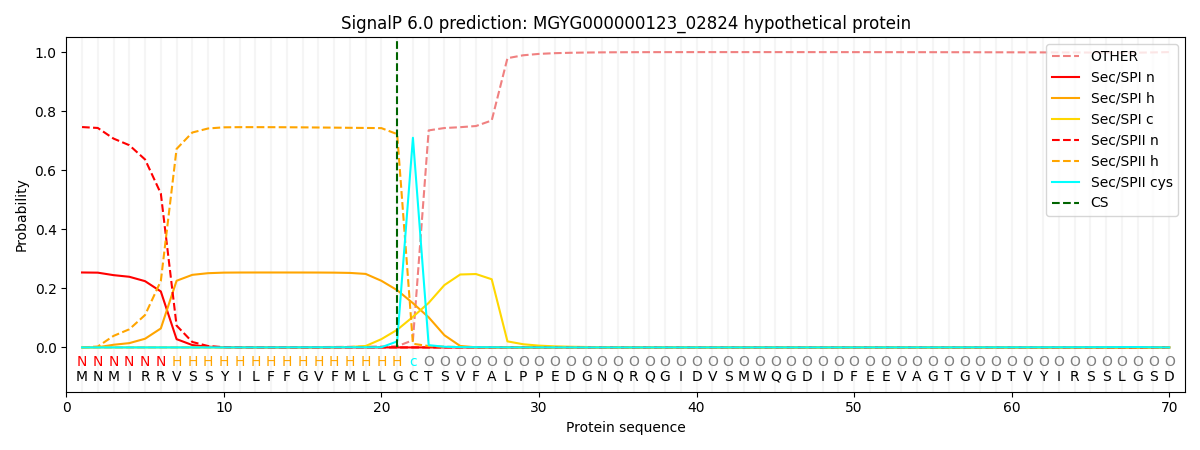
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000810 | 0.246593 | 0.752036 | 0.000210 | 0.000202 | 0.000152 |