You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000123_03675
You are here: Home > Sequence: MGYG000000123_03675
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
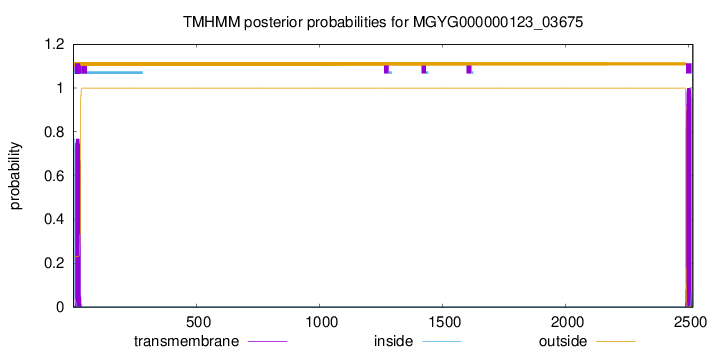
TMHMM annotations
Basic Information help
| Species | Blautia sp001304935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia; Blautia sp001304935 | |||||||||||
| CAZyme ID | MGYG000000123_03675 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 55751; End: 63304 Strand: - | |||||||||||
Full Sequence Download help
| MRSFKRGIAA ATTAALCLTS IPLGGLQLVS AADADRVIKL QPGLASTFHD TNEDGLGEFE | 60 |
| GWGTSLCWWA NRLGYSDKMT EEAARVFFSD EGLDMNIGRY NVGGGDLVGE PTKVPVNEKA | 120 |
| EFYDLETEGR KPEYKGKKMT VGTNTAMKSL KYTVSDADFG ITKGSEVGSF QSIGWINKLD | 180 |
| DEPGDGDNLR YTVKAEETGN YTVKLLLTLT GSNKRDVAIR VNDEKDYLVD ADTINSHIIA | 240 |
| SGNNNMLFLV TISDVALNAG DNTINIAGKS DWTLDFVKMA VIKSGEEGKL PEGEEFLHSE | 300 |
| HITRSDSAIP GYATDVTKIN ESEHELAWYE ENYDRADASC GYAWNYNWKA DENQINILKA | 360 |
| AAEASGEDFI AEAFSNSPPY FMTNSGCSSG AVDSNKDNLR TDSYRAFAVY MADVIEHWNK | 420 |
| EGVITFQSTT PMNEPYTSYW GAYSNKQEGC HFDQGESQSK IIEMLNEELK KREINIIICG | 480 |
| TDETSIDTAI TSYNALSDEA KGIIQRIDTH TYGGSRRAEL KELAEKENKN LWMSEVDGAY | 540 |
| TAGTDAGEMS AALGLAKQMM TDVNGLKANA WILWNAIDMH VDKEIKTSSD ADYASLEELY | 600 |
| NRVNMNSGYW GIAIGDHDNQ EILLTKKYYA YGQLSRYIRP GYTIIGSSDD TLAAYDPESG | 660 |
| KVVVVAVNTA GEDKTWKFDL SSFSTMGDNV TAIRTSGTLK DGENWADVSS EDTIAADTAK | 720 |
| KSITAVLKAN SITTYIVDGV TYDSSTEEIV RIDDVNVYTV EGVAAELPDT VEVVTNKNNT | 780 |
| LEKKVTWDLE GVDLTKDTKV TGTVEGTTMT ATANVQVVAP NMIYFIDCNS PESPKYAAMD | 840 |
| QYADLLNETA DQAYTEGSWG YLDEYGKYNG TVTDEYDTGW YANKDQVIKY TIPLEAGNYK | 900 |
| VNFGFKEWWN QQREMVITAE QDGKTVEIGK TNTWNGNNPW NTYSQDITIE NAGDVTFCVA | 960 |
| KQKNNDPVLS FIQIQKVLDL SELQDAITAA GEVDRAQYPA AKLAVLDAAV AAGRELMLKS | 1020 |
| ATTQDQIQEG AAAIQDAITG LGEGYTEEQI AANDYVLYLV NCGTSDASVV PDGYRRGLYQ | 1080 |
| SNVDQEYSAD AETGLSWGYE PNDENSKIVK GGSGAADITS SYIYMADTGV TFIKDVSGFK | 1140 |
| YKFEVPQRSN NDYVVTAGFK NPWDTRNVDI KLEGETVESD LKLTKGQLVE KSYKVEVHDG | 1200 |
| ELNVMVHSPK RTNQYGDPVL SYLMVEAVAA YTVEGLEAKI NSYEASMDGH SYSDATKKDF | 1260 |
| DLAVTEARKL IEEQSTDQKA IKNALNALEE AYNALKEVFT YSSITGTNGA QLFDNNGNKI | 1320 |
| QAHGGQIQQF TIDGETKYYW YGEDKTNGYR PVVGVHLYTS TDLYNWTDEG AVLKSIPVSE | 1380 |
| EDYGKDQEEG YEADLSIFET DEYFSSLYGD YKDQAPDDTE NYSSKLEEVY WNLAEDRTVI | 1440 |
| ERPKVLYNDA TGKYVMWFHA DGRTPTNTAD YGKARAGVAI SDSPTGPFKL LGTYKLHDSA | 1500 |
| DANHSWDNVG GAVRDMNLFK DDDGQAYVIY SSDGNLTTYI AKLNADYTGL ITDPADAVEG | 1560 |
| TGKEGEELTA SDYTRNFIDA SREAPAMFKY KDKYYIINSG CTGWSPNKAQ YAVADHPLGP | 1620 |
| WTVMGDPCVG DTKGTTFDTQ STCVFPVDAA NGKFIYMGDR WFNPDSGGDL SDSRYVWLPV | 1680 |
| EFLPGNQIQL KNYADWTLDE LENKASFEVT SELPTVVSSV SEIADVLPGT VMVDFGSGSV | 1740 |
| EKEVTWDIGS LDENKLGVAA VTGILTDGNR EFTHEISIVN PKLIYFFDSG ADTSEYYDTV | 1800 |
| NANLLGRLKN AVPDEAYTEE NGAGYVGVTN LVDKDKFDLG HHAGDTYLSN GWWAASGKDI | 1860 |
| VYAFDLEPGT YTVSAGFQEW WNTGRPSKMT ITSGDNLLAE KAFTVYNTST DLQVNQTFEV | 1920 |
| TEETKGKVTV TISKTGNPDP VISWIGVTHN DAAVNKDKLN TLITAADALK KDNYSEETWE | 1980 |
| ALSIILEEAR IASGNEEADQ KAIDEVTKAL AKALEDLEST ANKDALNNLI AEVDAMDNSI | 2040 |
| YTSASWAEYL SAVESARAEA QTVADNKAAL KDEIDAAVEA LRAAFYAGKE KLVTIQSVLE | 2100 |
| KGIAANTVPE SSKDSYTEES WADYAAALEA AQALIGSNDE QAVNDAIAAL EAAKNALEQK | 2160 |
| PEHVVDKTVL TEAIENAPSE EEAGLYTAKS WALYADVLTN AIDVRDSELA TQLDVDNAVT | 2220 |
| ALTQAQAELV TLEEKLAVMI EGYESHVSSA ESYTEESWTI YENALAAAKA LQNKPGLTEE | 2280 |
| EIDTADLALS EAYNGLTPVE VTVVDKGGLQ AVIEQAEQLN KEDYTEEAWK AFEESLNKAK | 2340 |
| AVYDNKDASQ KDVNKEITAL AEAMAELKKH PAKTEEETDK SALKALIDEA SQLKKDDYTG | 2400 |
| TTWKKFQKAF DDAANIYESE TATQEQIDAA TAALQAAMDQ LEKVTAGPNP KPQPPTDNNK | 2460 |
| DKGKNPSRPS GGGSQKGTSS AKTGDETPIG MFAGLGAVAL IAIFACTLSI LRKRKRS | 2517 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 300 | 735 | 3.6e-123 | 0.8574561403508771 |
| GH43 | 1335 | 1661 | 1.8e-97 | 0.9715447154471545 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd18825 | GH43_CtGH43-like | 8.09e-138 | 1320 | 1682 | 1 | 285 | Glycosyl hydrolase family 43 protein similar to Clostridium thermocellum exo-beta-1,3-galactanase CtGH43 and Ruminococcus champanellensis arabinanase Ara43A. This uncharacterized glycosyl hydrolase family 43 (GH43) subgroup belongs to a subgroup which includes characterized enzymes with exo-beta-1,3-galactanase (EC 3.2.1.145, also known as galactan 1,3-beta-galactosidase) activity such as Clostridium thermocellum (Ct1,3Gal43A or CtGH43) and Phanerochaete chrysosporium 1,3Gal43A (Pc1, 3Gal43A), and arabinanase (EC 3.2.1.99) activity such as Ruminococcus champanellensis Ara43A. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd08985 | GH43_CtGH43-like | 3.54e-93 | 1320 | 1682 | 1 | 273 | Glycosyl hydrolase family 43 protein such as Clostridium thermocellum exo-beta-1,3-galactanase CtGH43 and Ruminococcus champanellensis arabinanase Ara43A. This glycosyl hydrolase family 43 (GH43) subgroup includes characterized enzymes with exo-beta-1,3-galactanase (EC 3.2.1.145, also known as galactan 1,3-beta-galactosidase) activity such as Clostridium thermocellum (Ct1,3Gal43A or CtGH43) and Phanerochaete chrysosporium 1,3Gal43A (Pc1, 3Gal43A), and arabinanase (EC 3.2.1.99) activity such as Ruminococcus champanellensis Ara43A. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd18821 | GH43_Pc3Gal43A-like | 2.77e-84 | 1320 | 1682 | 1 | 262 | Glycosyl hydrolase family 43 protein such as Phanerochaete chrysosporium exo-beta-1,3-galactanase (Pc1, 3Gal43A, 1,3Gal43A). This glycosyl hydrolase family 43 (GH43) subgroup includes characterized enzymes with exo-beta-1,3-galactanase (EC 3.2.1.145, also known as galactan 1,3-beta-galactosidase) activity such as Phanerochaete chrysosporium 1,3Gal43A (Pc1, 3Gal43A), Fusarium oxysporum 12S Fo/1 (3Gal), and Streptomyces sp. 19(2012) SGalase1 and SGalase2. It belongs to the GH43_CtGH43 subgroup of the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43_CtGH43 includes proteins such as Clostridium thermocellum exo-beta-1,3-galactanase (Ct1,3Gal43A or CtGH43) which is comprised of the GH43 domain, a CBM13 domain, and a dockerin domain, exhibits an unusual ability to hydrolyze beta-1,3-galactan in the presence of a beta-1,6 linked branch, and is missing an essential acidic residue suggesting a mechanism by which it bypasses beta-1,6 linked branches in the substrate. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd18822 | GH43_CtGH43-like | 3.40e-82 | 1320 | 1682 | 1 | 266 | Glycosyl hydrolase family 43 protein such as Clostridium thermocellum exo-beta-1,3-galactanase (Ct1,3Gal43A or CtGH43). This glycosyl hydrolase family 43 (GH43) subgroup includes characterized enzymes with exo-beta-1,3-galactanase (EC 3.2.1.145, also known as galactan 1,3-beta-galactosidase) activity such as Clostridium thermocellum exo-beta-1,3-galactanase (Ct1,3Gal43A or CtGH43), Streptomyces avermitilis MA-4680 = NBRC 14893 (Sa1,3Gal43A;SAV2109) (1,3Gal43A), and Ruminiclostridium thermocellum ATCC 27405 (Ct1,3Gal43A;CtGH43;Cthe_0661) (1,3Gal43A). It belongs to the GH43_CtGH43 subgroup of the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43_CtGH43 includes proteins such as Clostridium thermocellum exo-beta-1,3-galactanase (Ct1,3Gal43A or CtGH43) which is comprised of the GH43 domain, a CBM13 domain, and a dockerin domain, exhibits an unusual ability to hydrolyze beta-1,3-galactan in the presence of a beta-1,6 linked branch, and is missing an essential acidic residue suggesting a mechanism by which it bypasses beta-1,6 linked branches in the substrate. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| cd18826 | GH43_CtGH43-like | 4.77e-75 | 1320 | 1682 | 1 | 269 | Glycosyl hydrolase family 43 protein similar to Clostridium thermocellum exo-beta-1,3-galactanase CtGH43 and Ruminococcus champanellensis arabinanase Ara43A. This uncharacterized glycosyl hydrolase family 43 (GH43) subgroup belongs to a subgroup which includes characterized enzymes with exo-beta-1,3-galactanase (EC 3.2.1.145, also known as galactan 1,3-beta-galactosidase) activity such as Clostridium thermocellum (Ct1,3Gal43A or CtGH43) and Phanerochaete chrysosporium 1,3Gal43A (Pc1, 3Gal43A), and arabinanase (EC 3.2.1.99) activity such as Ruminococcus champanellensis Ara43A. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBE99233.1 | 0.0 | 1 | 2516 | 1 | 2521 |
| ASU27677.1 | 0.0 | 1 | 2517 | 1 | 2523 |
| ANU74869.1 | 0.0 | 1 | 2517 | 1 | 2523 |
| QQQ92416.1 | 0.0 | 1 | 2517 | 1 | 2523 |
| QJU15030.1 | 0.0 | 1 | 2517 | 1 | 2523 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EUG_A | 3.16e-61 | 1314 | 1699 | 37 | 374 | TheGH43, Beta 1,3 Galactosidase, BT3683 with galactoimidazole [Bacteroides thetaiotaomicron],6EUH_A The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin [Bacteroides thetaiotaomicron VPI-5482],6EUH_B The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin [Bacteroides thetaiotaomicron VPI-5482],6EUH_C The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin [Bacteroides thetaiotaomicron VPI-5482],6EUI_A The GH43, Beta 1,3 Galactosidase, BT3683 with galactose [Bacteroides thetaiotaomicron VPI-5482] |
| 3VSF_A | 1.09e-48 | 1308 | 1731 | 44 | 381 | ChainA, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSF_B Chain B, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSF_C Chain C, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSF_D Chain D, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSF_E Chain E, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSF_F Chain F, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSZ_A Chain A, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSZ_B Chain B, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSZ_C Chain C, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSZ_D Chain D, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSZ_E Chain E, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VSZ_F Chain F, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT0_A Chain A, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT0_B Chain B, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT0_C Chain C, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT0_D Chain D, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT0_E Chain E, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT0_F Chain F, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT1_A Chain A, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT1_B Chain B, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT1_C Chain C, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT1_D Chain D, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT1_E Chain E, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT1_F Chain F, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT2_A Chain A, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT2_B Chain B, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT2_C Chain C, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT2_D Chain D, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT2_E Chain E, Ricin B lectin [Acetivibrio thermocellus ATCC 27405],3VT2_F Chain F, Ricin B lectin [Acetivibrio thermocellus ATCC 27405] |
| 6EUF_A | 2.12e-43 | 1314 | 1713 | 13 | 317 | TheGH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482],6EUF_B The GH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482],6EUF_C The GH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482],6EUF_D The GH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482],6EUJ_A The GH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482],6EUJ_B The GH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482],6EUJ_C The GH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482],6EUJ_D The GH43, Beta 1,3 Galactosidase, BT0265 [Bacteroides thetaiotaomicron VPI-5482] |
| 7BYS_A | 5.41e-39 | 1308 | 1685 | 5 | 280 | ChainA, Galactan 1,3-beta-galactosidase [Phanerodontia chrysosporium],7BYS_B Chain B, Galactan 1,3-beta-galactosidase [Phanerodontia chrysosporium],7BYT_A Chain A, Galactan 1,3-beta-galactosidase [Phanerodontia chrysosporium] |
| 7BYV_A | 1.36e-38 | 1308 | 1685 | 6 | 281 | ChainA, Galactan 1,3-beta-galactosidase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q76FP5 | 2.30e-45 | 345 | 693 | 125 | 433 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
| A0A401ETL2 | 2.02e-26 | 351 | 741 | 172 | 641 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| E8MGH9 | 1.30e-15 | 2165 | 2363 | 1666 | 1871 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
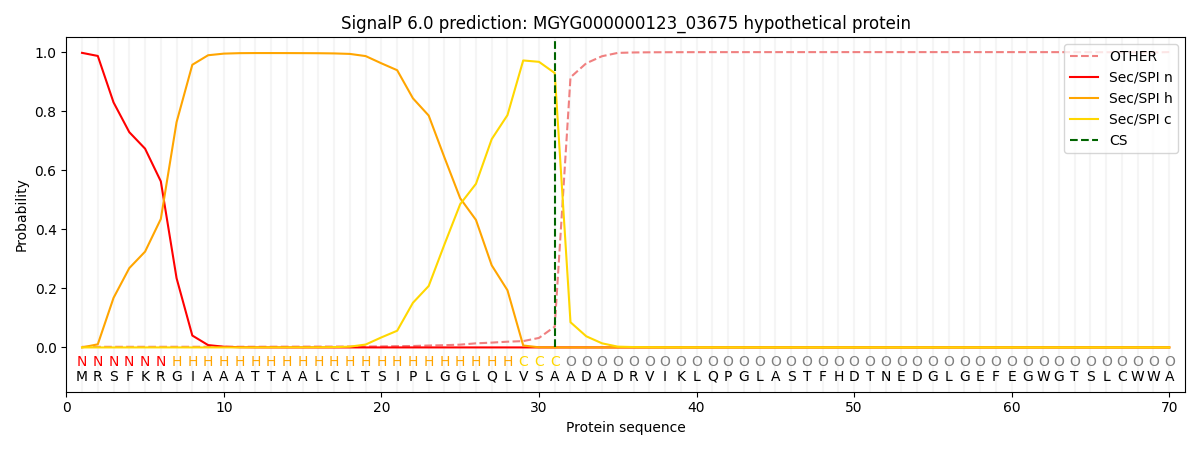
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003605 | 0.994484 | 0.000268 | 0.001021 | 0.000323 | 0.000260 |