You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000123_05102
You are here: Home > Sequence: MGYG000000123_05102
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
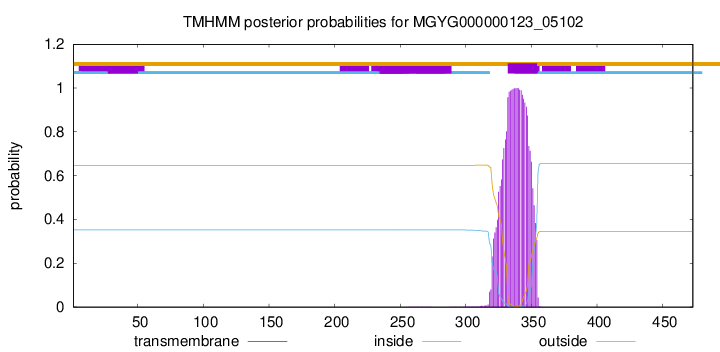
TMHMM annotations
Basic Information help
| Species | Blautia sp001304935 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Blautia; Blautia sp001304935 | |||||||||||
| CAZyme ID | MGYG000000123_05102 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 91; End: 1512 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13702 | Lysozyme_like | 2.46e-35 | 368 | 468 | 1 | 92 | Lysozyme-like. |
| cd16891 | CwlT-like | 4.59e-33 | 374 | 468 | 1 | 86 | CwlT-like N-terminal lysozyme domain and similar domains. CwlT is a bifunctional cell wall hydrolase containing an N-terminal lysozyme domain and a C-terminal NlpC/P60 endopeptidase domain (gamma-d-D-glutamyl-L-diamino acid endopeptidase), and has been implicated in the spread of transposons. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| cd00254 | LT-like | 0.001 | 392 | 468 | 2 | 79 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJU17139.1 | 5.05e-263 | 1 | 468 | 1 | 478 |
| QRT48860.1 | 1.59e-41 | 351 | 468 | 218 | 335 |
| CBL18457.1 | 4.05e-41 | 346 | 468 | 37 | 159 |
| QHB23689.1 | 1.33e-40 | 364 | 468 | 275 | 379 |
| ASM68260.1 | 1.33e-40 | 364 | 468 | 275 | 379 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96645 | 7.00e-08 | 363 | 468 | 49 | 142 | Probable endopeptidase YddH OS=Bacillus subtilis (strain 168) OX=224308 GN=yddH PE=3 SV=1 |
| O34636 | 3.38e-06 | 367 | 468 | 48 | 140 | Uncharacterized membrane protein YocA OS=Bacillus subtilis (strain 168) OX=224308 GN=yocA PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
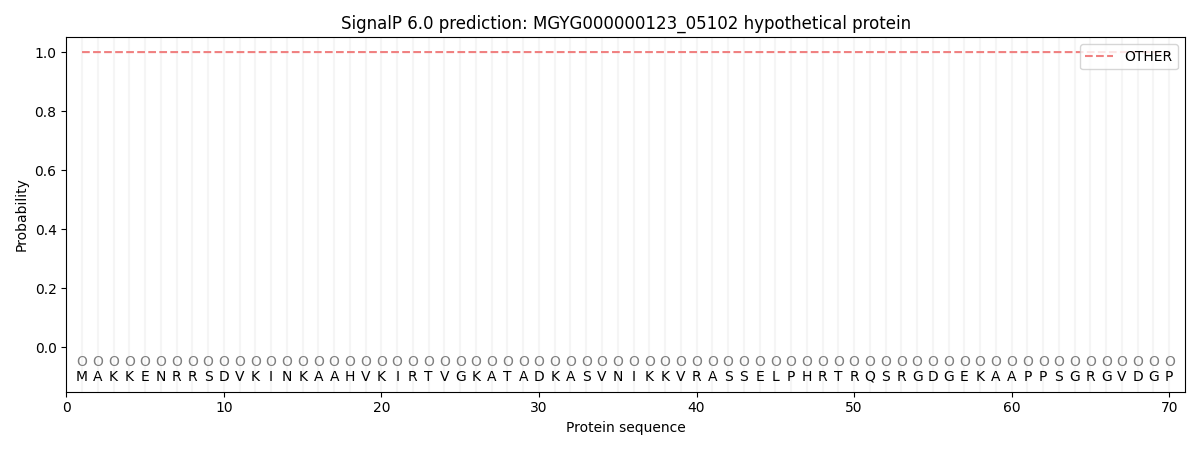
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000068 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |