You are browsing environment: HUMAN GUT
MGYG000000128_00949
Basic Information
help
Species
Alistipes sp902363575
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp902363575
CAZyme ID
MGYG000000128_00949
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000128
2906288
Isolate
Canada
North America
Gene Location
Start: 195086;
End: 196717
Strand: +
No EC number prediction in MGYG000000128_00949.
Family
Start
End
Evalue
family coverage
GT83
4
426
1.2e-63
0.7944444444444444
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
2.41e-18
18
342
22
354
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
5EZM_A
1.54e-07
23
269
51
300
CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
Q3KCC9
6.46e-11
8
317
28
337
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT1 PE=3 SV=1
more
A8FRR0
1.35e-09
23
317
26
319
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Shewanella sediminis (strain HAW-EB3) OX=425104 GN=arnT PE=3 SV=1
more
O67270
3.57e-08
30
327
29
318
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1
more
A0KGY4
1.08e-06
1
323
1
323
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=arnT PE=3 SV=1
more
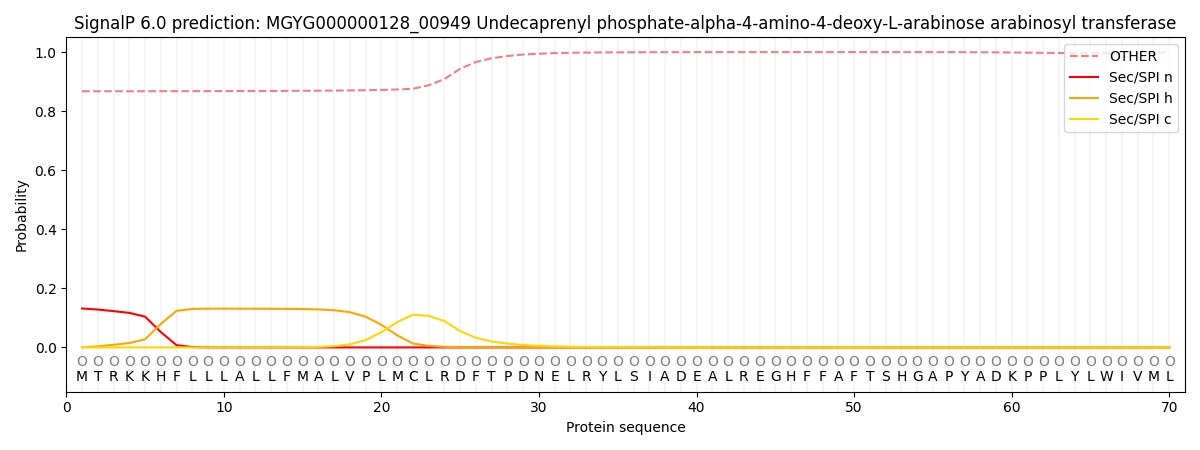
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.871952
0.125340
0.001440
0.000303
0.000258
0.000716
start
end
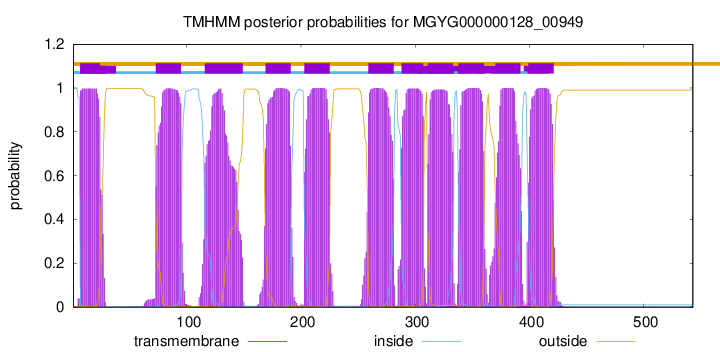
7
24
73
95
116
149
169
191
203
225
259
281
288
307
311
333
338
360
370
392
399
421