You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000132_01536
You are here: Home > Sequence: MGYG000000132_01536
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
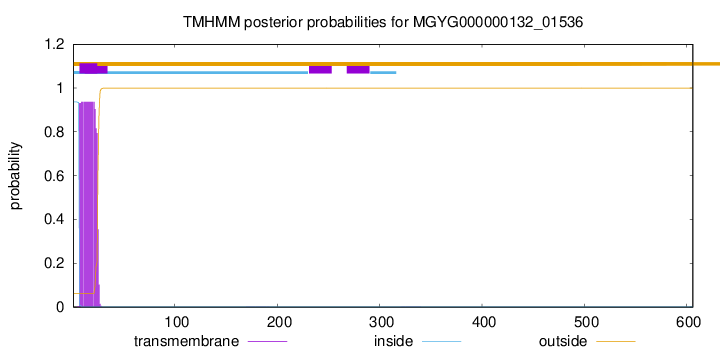
TMHMM annotations
Basic Information help
| Species | Beduini sp902363625 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Beduini; Beduini sp902363625 | |||||||||||
| CAZyme ID | MGYG000000132_01536 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 69911; End: 71731 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 312 | 426 | 5.5e-19 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14256 | Dockerin_I | 4.10e-16 | 548 | 603 | 1 | 56 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| COG4193 | LytD | 4.18e-15 | 248 | 427 | 37 | 228 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam00404 | Dockerin_1 | 2.81e-13 | 549 | 604 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| pfam01832 | Glucosaminidase | 2.17e-09 | 312 | 377 | 1 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| COG1705 | FlgJ | 5.65e-05 | 288 | 393 | 19 | 151 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM11023.1 | 8.72e-131 | 11 | 581 | 16 | 607 |
| QUN13370.1 | 5.88e-113 | 28 | 577 | 31 | 586 |
| BCT44532.1 | 1.21e-97 | 45 | 583 | 50 | 610 |
| QSI26463.1 | 1.49e-88 | 62 | 583 | 31 | 562 |
| QJA02388.1 | 2.50e-86 | 115 | 583 | 81 | 564 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 1.15e-13 | 301 | 427 | 161 | 282 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 3.71e-14 | 165 | 427 | 424 | 650 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 3.92e-14 | 165 | 427 | 468 | 694 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.017654 | 0.818096 | 0.162904 | 0.000672 | 0.000344 | 0.000294 |