You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000134_01119
You are here: Home > Sequence: MGYG000000134_01119
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Longibaculum muris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Longibaculum; Longibaculum muris | |||||||||||
| CAZyme ID | MGYG000000134_01119 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 163014; End: 168215 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 1134 | 1250 | 9.6e-24 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.59e-18 | 1132 | 1248 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 5.31e-07 | 1135 | 1252 | 16 | 142 | Substituted updates: Jan 31, 2002 |
| COG1196 | Smc | 6.51e-07 | 1412 | 1694 | 739 | 1021 | Chromosome segregation ATPase [Cell cycle control, cell division, chromosome partitioning]. |
| COG0419 | SbcC | 9.00e-07 | 1425 | 1683 | 491 | 728 | DNA repair exonuclease SbcCD ATPase subunit [Replication, recombination and repair]. |
| COG1340 | COG1340 | 1.09e-04 | 1429 | 1683 | 1 | 274 | Uncharacterized coiled-coil protein, contains DUF342 domain [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQY26898.1 | 0.0 | 6 | 1686 | 17 | 1712 |
| QQV06122.1 | 0.0 | 6 | 1686 | 17 | 1712 |
| QPS14238.1 | 0.0 | 6 | 1686 | 17 | 1712 |
| QMW75427.1 | 0.0 | 6 | 1686 | 6 | 1701 |
| QQY26901.1 | 2.38e-62 | 37 | 557 | 499 | 953 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RV9_A | 2.16e-17 | 1126 | 1252 | 7 | 135 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 2.22e-17 | 1126 | 1252 | 8 | 136 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 4.13e-17 | 1126 | 1252 | 8 | 136 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 2RVA_A | 1.70e-15 | 1126 | 1252 | 7 | 136 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZZ5_A | 1.75e-15 | 1126 | 1252 | 8 | 137 | X-raycrystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ5_B X-ray crystal structure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_A X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZZ8_B X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
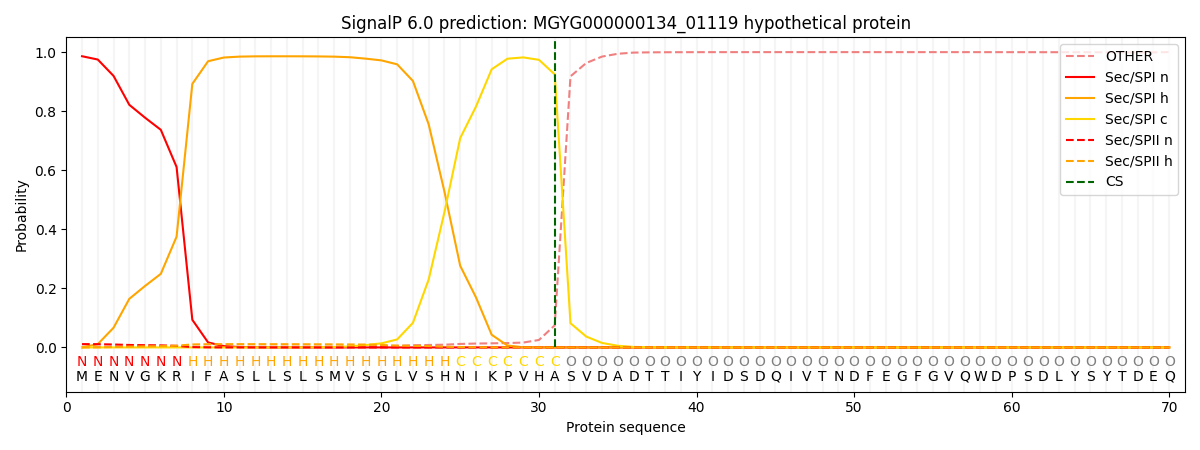
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004244 | 0.981550 | 0.012360 | 0.001245 | 0.000306 | 0.000251 |
