You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000135_03750
You are here: Home > Sequence: MGYG000000135_03750
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
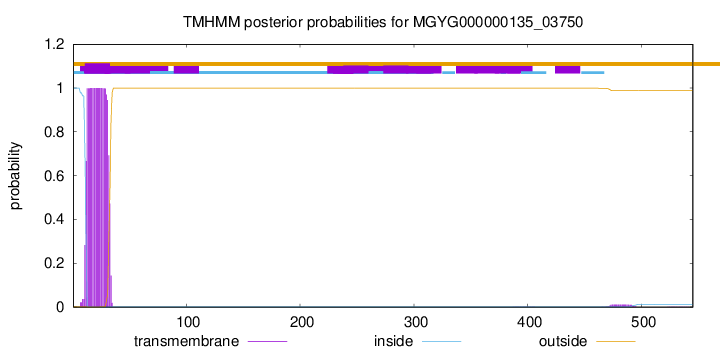
TMHMM annotations
Basic Information help
| Species | Murimonas intestini | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Murimonas; Murimonas intestini | |||||||||||
| CAZyme ID | MGYG000000135_03750 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | Thiol-disulfide oxidoreductase ResA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 226399; End: 228036 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 218 | 533 | 3e-82 | 0.9844236760124611 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02966 | TlpA_like_family | 2.60e-15 | 76 | 190 | 3 | 112 | TlpA-like family; composed of TlpA, ResA, DsbE and similar proteins. TlpA, ResA and DsbE are bacterial protein disulfide reductases with important roles in cytochrome maturation. They are membrane-anchored proteins with a soluble TRX domain containing a CXXC motif located in the periplasm. The TRX domains of this family contain an insert, approximately 25 residues in length, which correspond to an extra alpha helix and a beta strand when compared with TRX. TlpA catalyzes an essential reaction in the biogenesis of cytochrome aa3, while ResA and DsbE are essential proteins in cytochrome c maturation. Also included in this family are proteins containing a TlpA-like TRX domain with domain architectures similar to E. coli DipZ protein, and the N-terminal TRX domain of PilB protein from Neisseria which acts as a disulfide reductase that can recylce methionine sulfoxide reductases. |
| pfam00578 | AhpC-TSA | 1.38e-10 | 69 | 191 | 2 | 124 | AhpC/TSA family. This family contains proteins related to alkyl hydroperoxide reductase (AhpC) and thiol specific antioxidant (TSA). |
| PRK03147 | PRK03147 | 5.87e-10 | 69 | 188 | 38 | 151 | thiol-disulfide oxidoreductase ResA. |
| pfam08534 | Redoxin | 3.28e-08 | 69 | 209 | 3 | 147 | Redoxin. This family of redoxins includes peroxiredoxin, thioredoxin and glutaredoxin proteins. |
| pfam13905 | Thioredoxin_8 | 2.56e-07 | 93 | 188 | 1 | 94 | Thioredoxin-like. Thioredoxins are small enzymes that participate in redox reactions, via the reversible oxidation of an active centre disulfide bond. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCN29944.1 | 1.71e-104 | 35 | 535 | 45 | 538 |
| APF21752.1 | 2.04e-83 | 52 | 535 | 41 | 516 |
| QSX01740.1 | 7.93e-83 | 21 | 535 | 8 | 516 |
| QUF82251.1 | 7.93e-83 | 21 | 535 | 8 | 516 |
| QJU43754.1 | 7.93e-83 | 21 | 535 | 8 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 3.90e-41 | 213 | 540 | 18 | 350 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
| 4NMU_A | 1.69e-09 | 69 | 188 | 12 | 125 | CrystalStructure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis],4NMU_B Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis],4NMU_C Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis],4NMU_D Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' [Bacillus anthracis] |
| 3C73_A | 2.33e-08 | 79 | 198 | 10 | 124 | ChainA, Thiol-disulfide oxidoreductase resA [unidentified],3C73_B Chain B, Thiol-disulfide oxidoreductase resA [unidentified] |
| 3C71_A | 2.48e-08 | 69 | 198 | 3 | 127 | ChainA, Thiol-disulfide oxidoreductase resA [Bacillus subtilis] |
| 1ST9_A | 6.25e-08 | 69 | 198 | 3 | 127 | CrystalStructure of a Soluble Domain of ResA in the Oxidised Form [Bacillus subtilis],1ST9_B Crystal Structure of a Soluble Domain of ResA in the Oxidised Form [Bacillus subtilis],1SU9_A Reduced structure of the soluble domain of ResA [Bacillus subtilis],1SU9_B Reduced structure of the soluble domain of ResA [Bacillus subtilis],2H1D_A ResA pH 9.25 [Bacillus subtilis],2H1D_B ResA pH 9.25 [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 9.63e-26 | 215 | 533 | 42 | 350 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
| Q73B22 | 1.82e-09 | 69 | 188 | 38 | 151 | Thiol-disulfide oxidoreductase ResA OS=Bacillus cereus (strain ATCC 10987 / NRS 248) OX=222523 GN=resA PE=3 SV=1 |
| Q6HL81 | 1.82e-09 | 69 | 188 | 38 | 151 | Thiol-disulfide oxidoreductase ResA OS=Bacillus thuringiensis subsp. konkukian (strain 97-27) OX=281309 GN=resA PE=3 SV=1 |
| Q81SZ9 | 4.56e-09 | 66 | 188 | 35 | 151 | Thiol-disulfide oxidoreductase ResA OS=Bacillus anthracis OX=1392 GN=resA PE=1 SV=1 |
| A0RBT0 | 4.56e-09 | 66 | 188 | 35 | 151 | Thiol-disulfide oxidoreductase ResA OS=Bacillus thuringiensis (strain Al Hakam) OX=412694 GN=resA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
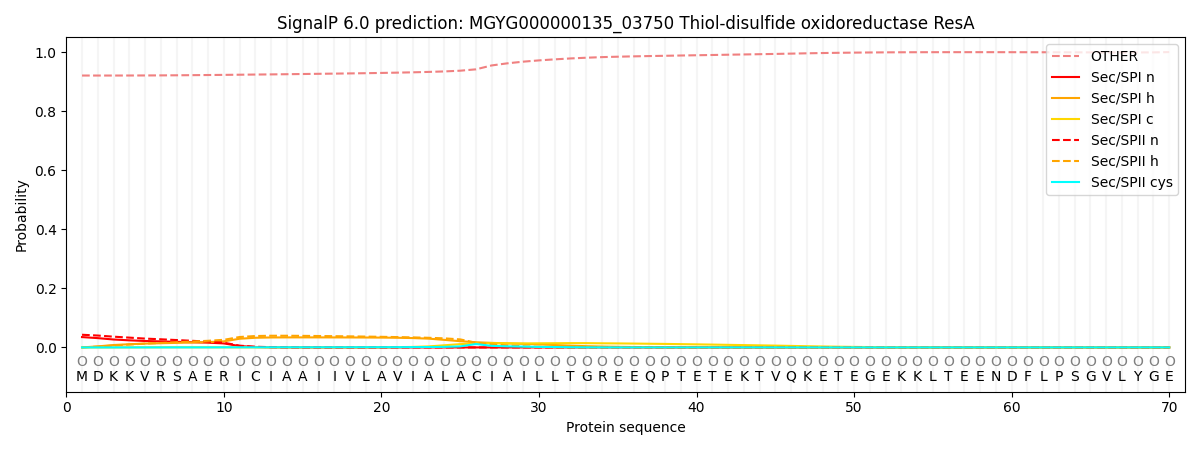
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.922280 | 0.032297 | 0.043265 | 0.000205 | 0.000138 | 0.001834 |