You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000136_00867
You are here: Home > Sequence: MGYG000000136_00867
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
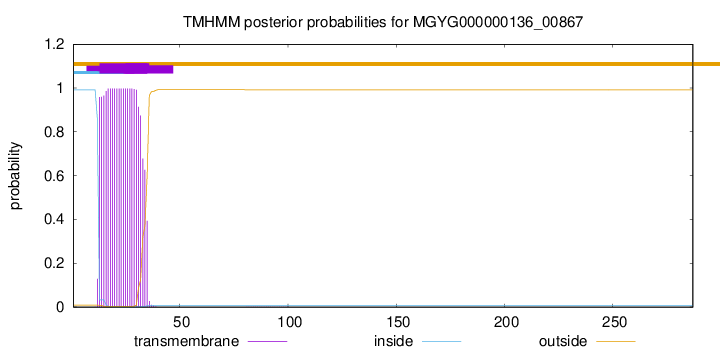
TMHMM annotations
Basic Information help
| Species | Agathobacter sp000434275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter; Agathobacter sp000434275 | |||||||||||
| CAZyme ID | MGYG000000136_00867 | |||||||||||
| CAZy Family | GH166 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16689; End: 17552 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH166 | 65 | 282 | 1.7e-59 | 0.9821428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3868 | COG3868 | 2.67e-22 | 60 | 287 | 77 | 305 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
| pfam03537 | Glyco_hydro_114 | 2.18e-20 | 66 | 159 | 20 | 113 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG2342 | COG2342 | 3.62e-18 | 42 | 259 | 24 | 254 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL08521.1 | 5.36e-109 | 17 | 285 | 22 | 282 |
| VCV23723.1 | 2.17e-108 | 17 | 285 | 22 | 282 |
| CBL11057.1 | 2.17e-108 | 17 | 285 | 22 | 282 |
| AOZ95737.1 | 9.34e-96 | 20 | 285 | 22 | 285 |
| QHI71547.1 | 5.11e-87 | 17 | 285 | 4 | 271 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
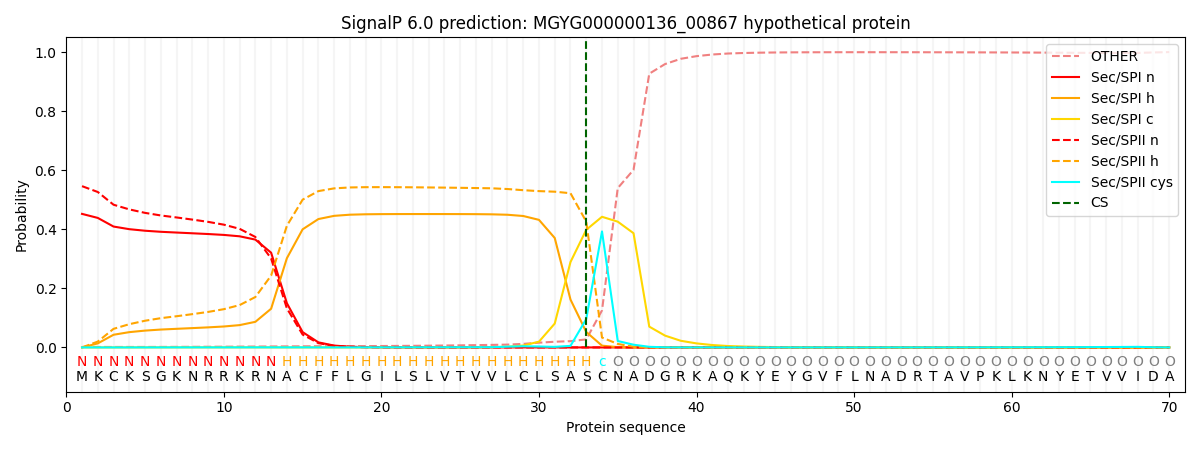
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002711 | 0.442534 | 0.553854 | 0.000404 | 0.000252 | 0.000234 |