You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000136_01153
You are here: Home > Sequence: MGYG000000136_01153
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Agathobacter sp000434275 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter; Agathobacter sp000434275 | |||||||||||
| CAZyme ID | MGYG000000136_01153 | |||||||||||
| CAZy Family | CBM86 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 98877; End: 102953 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 380 | 768 | 2.1e-81 | 0.9636963696369637 |
| CBM9 | 927 | 1098 | 1.1e-49 | 0.9945054945054945 |
| CBM86 | 52 | 161 | 1.3e-34 | 0.9906542056074766 |
| CBM22 | 178 | 328 | 5.5e-19 | 0.9618320610687023 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00005 | CBM9_like_1 | 2.57e-75 | 922 | 1099 | 6 | 185 | DOMON-like type 9 carbohydrate binding module of xylanases. Family 9 carbohydrate-binding modules (CBM9) play a role in the microbial degradation of cellulose and hemicellulose (materials found in plants). The domain has previously been called cellulose-binding domain. The polysaccharide binding sites of CBMs with available 3D structure have been found to be either flat surfaces with interactions formed by predominantly aromatic residues (tryptophan and tyrosine), or extended shallow grooves. The CBM9 domain frequently occurs in tandem repeats; members found in this subfamily typically co-occur with glycosyl hydrolase family 10 domains and are annotated as endo-1,4-beta-xylanases. CBM9 from Thermotoga maritima xylanase 10A is reported to have specificity for polysaccharide reducing ends. |
| pfam00331 | Glyco_hydro_10 | 6.95e-68 | 384 | 767 | 13 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 8.77e-68 | 451 | 764 | 12 | 262 | Glycosyl hydrolase family 10. |
| pfam06452 | CBM9_1 | 1.98e-56 | 927 | 1099 | 1 | 182 | Carbohydrate family 9 binding domain-like. CBM9_1 is a C-terminal domain on bacterial xylanase proteins, and it is tandemly repeated in a number of family-members. The CBM9 module binds to amorphous and crystalline cellulose and a range of soluble di- and monosaccharides as well as to cello- and xylo- oligomers of different degrees of polymerization. Comparison of the glucose and cellobiose complexes during crystallisation reveals surprising differences in binding of these two substrates by CBM9-2. Cellobiose was found to bind in a distinct orientation from glucose, while still maintaining optimal stacking and electrostatic interactions with the reducing end sugar. |
| cd20907 | CBM86 | 2.88e-47 | 40 | 166 | 1 | 127 | carbohydrate binding module family 86. This family describes what is most likely a xylan-binding module such as found in the Xyn10A protein of Roseburia intestinalis L1-82, which is involved in the extracellular capture and breakdown of xylan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VCV24088.1 | 0.0 | 2 | 1098 | 5 | 1101 |
| CBL13458.1 | 0.0 | 14 | 1098 | 1 | 1085 |
| EEV01588.1 | 0.0 | 14 | 1098 | 1 | 1085 |
| QJU17095.1 | 7.59e-281 | 176 | 1099 | 230 | 1147 |
| BAA09971.1 | 1.00e-244 | 177 | 1096 | 44 | 823 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SGF_A | 3.21e-49 | 42 | 170 | 7 | 135 | Molecularinsight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. [Roseburia intestinalis L1-82],6SGF_B Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. [Roseburia intestinalis L1-82],6SGF_C Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. [Roseburia intestinalis L1-82],6SGF_D Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. [Roseburia intestinalis L1-82],6SGF_E Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. [Roseburia intestinalis L1-82],6SGF_F Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. [Roseburia intestinalis L1-82] |
| 2W5F_A | 9.32e-48 | 213 | 762 | 53 | 521 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 2WYS_A | 1.10e-45 | 213 | 762 | 53 | 521 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WYS_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_A Chain A, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2WZE_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 6D5C_A | 1.30e-45 | 367 | 768 | 16 | 350 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 5OFJ_A | 4.51e-45 | 366 | 769 | 3 | 339 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P36917 | 1.11e-100 | 237 | 1101 | 246 | 1045 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| P38535 | 2.29e-95 | 178 | 1101 | 38 | 897 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| Q60042 | 6.50e-95 | 302 | 1098 | 301 | 1053 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| Q60037 | 4.42e-94 | 302 | 1098 | 305 | 1057 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| O69230 | 7.99e-86 | 206 | 1098 | 217 | 1083 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
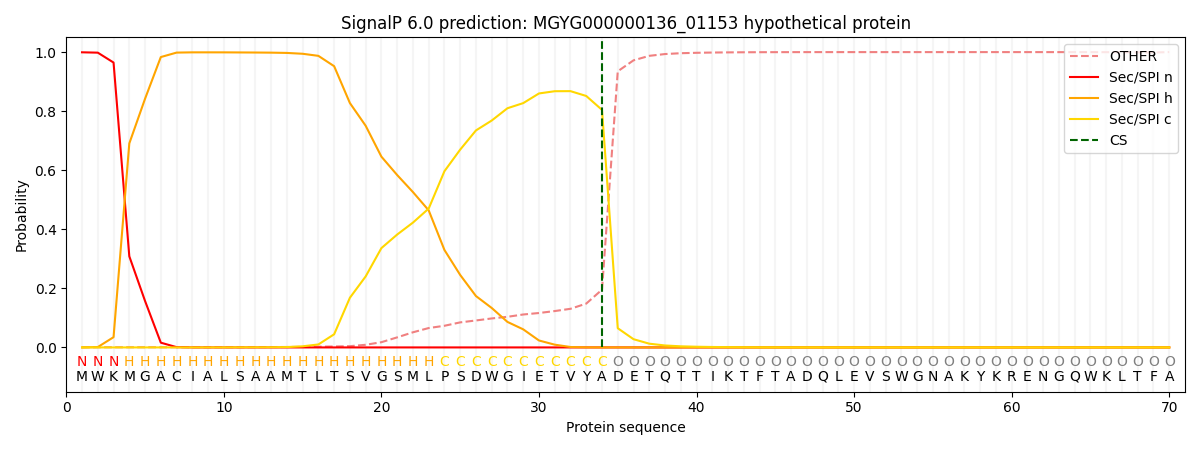
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002278 | 0.996434 | 0.000346 | 0.000389 | 0.000289 | 0.000253 |
