You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000137_02453
You are here: Home > Sequence: MGYG000000137_02453
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
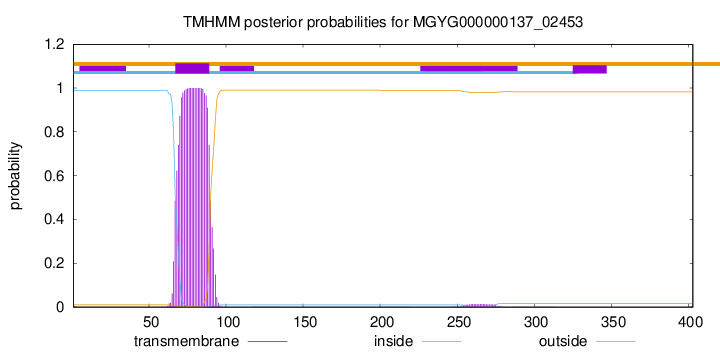
TMHMM annotations
Basic Information help
| Species | UBA7160 sp902363665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UBA7160; UBA7160 sp902363665 | |||||||||||
| CAZyme ID | MGYG000000137_02453 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14807; End: 16018 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16891 | CwlT-like | 1.71e-56 | 110 | 255 | 1 | 151 | CwlT-like N-terminal lysozyme domain and similar domains. CwlT is a bifunctional cell wall hydrolase containing an N-terminal lysozyme domain and a C-terminal NlpC/P60 endopeptidase domain (gamma-d-D-glutamyl-L-diamino acid endopeptidase), and has been implicated in the spread of transposons. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| pfam13702 | Lysozyme_like | 2.51e-56 | 104 | 254 | 1 | 165 | Lysozyme-like. |
| pfam05257 | CHAP | 1.23e-13 | 290 | 380 | 1 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
| pfam01464 | SLT | 0.002 | 114 | 224 | 1 | 113 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK79810.1 | 5.88e-250 | 2 | 403 | 257 | 658 |
| QCU01783.1 | 9.39e-248 | 2 | 403 | 242 | 643 |
| CBK88512.1 | 7.19e-246 | 1 | 403 | 245 | 647 |
| QIB55652.1 | 3.63e-175 | 96 | 403 | 299 | 605 |
| QMW76485.1 | 3.63e-175 | 96 | 403 | 299 | 605 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HPE_A | 6.09e-15 | 103 | 253 | 20 | 176 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 4FDY_A | 2.86e-14 | 103 | 253 | 22 | 177 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96645 | 4.05e-31 | 103 | 365 | 53 | 293 | Probable endopeptidase YddH OS=Bacillus subtilis (strain 168) OX=224308 GN=yddH PE=3 SV=1 |
| O34636 | 1.84e-10 | 105 | 232 | 50 | 168 | Uncharacterized membrane protein YocA OS=Bacillus subtilis (strain 168) OX=224308 GN=yocA PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
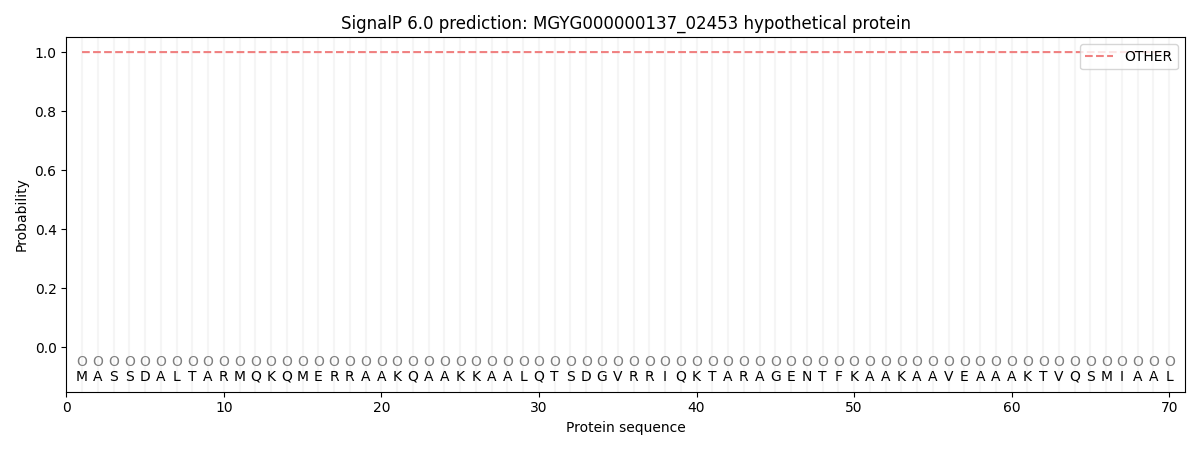
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000007 | 0.000048 | 0.000002 | 0.000000 | 0.000000 | 0.000000 |