You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000138_01391
You are here: Home > Sequence: MGYG000000138_01391
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides johnsonii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides johnsonii | |||||||||||
| CAZyme ID | MGYG000000138_01391 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 385321; End: 388200 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 27 | 717 | 5e-70 | 0.7021276595744681 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.09e-26 | 22 | 451 | 3 | 426 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 9.91e-15 | 28 | 449 | 9 | 440 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 3.75e-12 | 125 | 431 | 141 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02836 | Glyco_hydro_2_C | 2.30e-09 | 310 | 450 | 1 | 154 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam00703 | Glyco_hydro_2 | 6.80e-09 | 204 | 308 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50289.1 | 0.0 | 1 | 959 | 1 | 959 |
| QKH97806.1 | 0.0 | 23 | 956 | 17 | 937 |
| QUT54529.1 | 0.0 | 23 | 956 | 17 | 937 |
| QUR50384.1 | 0.0 | 23 | 956 | 17 | 937 |
| ABR43180.1 | 0.0 | 23 | 956 | 17 | 937 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZJV_A | 6.61e-17 | 31 | 457 | 57 | 468 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A [Arthrobacter sp. 32cB],6ZJW_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with galactose [Arthrobacter sp. 32cB],6ZJX_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose [Arthrobacter sp. 32cB] |
| 6ETZ_A | 8.58e-17 | 31 | 457 | 36 | 447 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB [Arthrobacter sp. 32cB],6H1P_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB - data collected at room temperature [Arthrobacter sp. 32cB] |
| 6SEB_A | 8.69e-17 | 31 | 457 | 57 | 468 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG [Arthrobacter sp. 32cB],6SEC_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cBon complex with ONPG [Arthrobacter sp. 32cB],6SED_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose [Arthrobacter sp. 32cB] |
| 6ZJP_A | 8.69e-17 | 31 | 457 | 57 | 468 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q [Arthrobacter sp. 32cB] |
| 6ZJQ_A | 8.70e-17 | 31 | 457 | 59 | 470 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with galactose [Arthrobacter sp. 32cB],6ZJR_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with lactulose [Arthrobacter sp. 32cB] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O52847 | 2.12e-16 | 98 | 453 | 137 | 503 | Beta-galactosidase OS=Priestia megaterium (strain DSM 319 / IMG 1521) OX=592022 GN=bgaM PE=3 SV=1 |
| Q59140 | 2.18e-14 | 32 | 457 | 30 | 461 | Beta-galactosidase OS=Arthrobacter sp. (strain B7) OX=86041 GN=lacZ PE=1 SV=1 |
| A4W7D2 | 2.55e-13 | 33 | 431 | 55 | 463 | Beta-galactosidase OS=Enterobacter sp. (strain 638) OX=399742 GN=lacZ PE=3 SV=1 |
| Q47077 | 2.96e-12 | 31 | 431 | 53 | 463 | Beta-galactosidase OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| Q2XQU3 | 2.96e-12 | 30 | 431 | 52 | 463 | Beta-galactosidase 2 OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
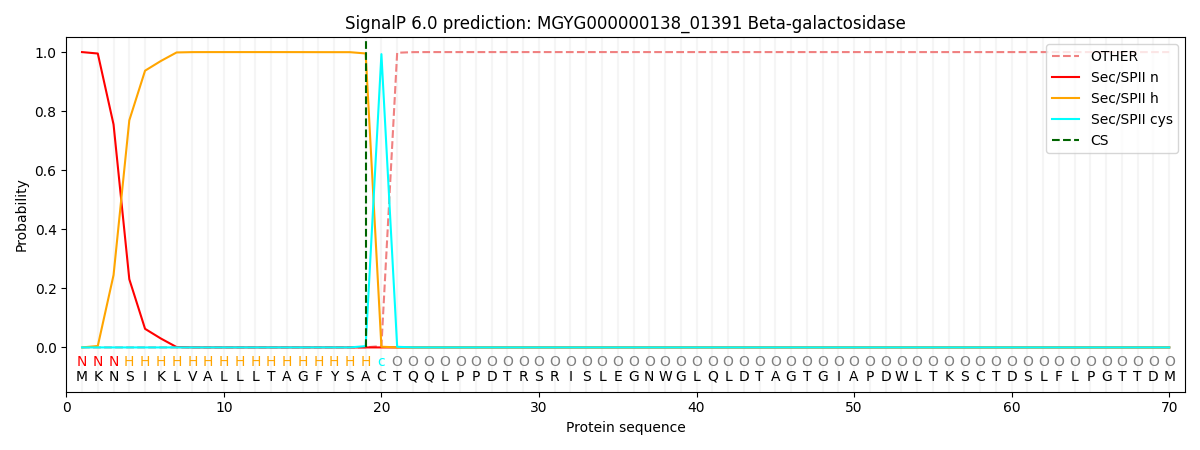
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000044 | 0.000000 | 0.000000 | 0.000000 |
