You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000138_03900
You are here: Home > Sequence: MGYG000000138_03900
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
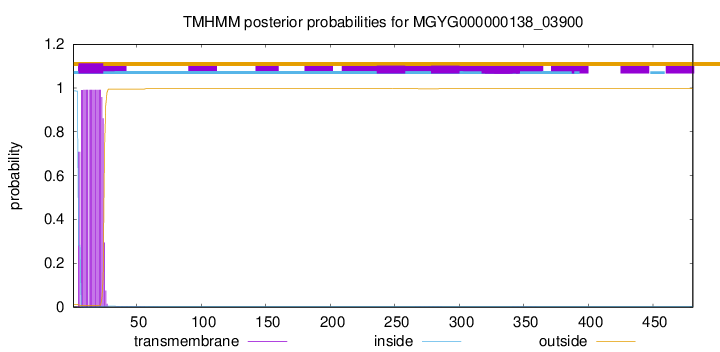
TMHMM annotations
Basic Information help
| Species | Parabacteroides johnsonii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides johnsonii | |||||||||||
| CAZyme ID | MGYG000000138_03900 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47185; End: 48630 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 34 | 222 | 8.1e-44 | 0.9845360824742269 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0657 | Aes | 2.32e-39 | 205 | 463 | 21 | 298 | Acetyl esterase/lipase [Lipid transport and metabolism]. |
| pfam07859 | Abhydrolase_3 | 5.32e-32 | 268 | 451 | 1 | 205 | alpha/beta hydrolase fold. This catalytic domain is found in a very wide range of enzymes. |
| cd01833 | XynB_like | 4.06e-29 | 32 | 222 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| COG1506 | DAP2 | 1.55e-22 | 230 | 477 | 360 | 615 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| pfam13472 | Lipase_GDSL_2 | 1.50e-18 | 37 | 213 | 2 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAX78799.1 | 6.09e-180 | 25 | 481 | 31 | 494 |
| ATC65549.1 | 1.88e-138 | 34 | 481 | 40 | 492 |
| QOV88633.1 | 5.94e-64 | 34 | 226 | 32 | 226 |
| QDT00894.1 | 9.49e-61 | 235 | 481 | 32 | 283 |
| QNN20991.1 | 8.91e-59 | 26 | 226 | 26 | 226 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AO9_A | 2.00e-52 | 240 | 481 | 20 | 278 | Thestructure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-native [Thermogutta terrifontis],5AOA_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-Propionate bound [Thermogutta terrifontis],5AOB_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-butyrate bound [Thermogutta terrifontis],5AOC_A The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound [Thermogutta terrifontis] |
| 7BFN_A | 2.06e-52 | 240 | 481 | 21 | 279 | ChainA, Esterase [Thermogutta terrifontis] |
| 7BFO_A | 1.11e-51 | 240 | 481 | 21 | 279 | ChainA, Esterase [Thermogutta terrifontis],7BFR_A Chain A, Esterase [Thermogutta terrifontis],7BFT_A Chain A, Esterase [Thermogutta terrifontis],7BFU_A Chain A, Esterase [Thermogutta terrifontis],7BFV_A Chain A, Esterase [Thermogutta terrifontis] |
| 4Q3K_A | 3.85e-17 | 263 | 473 | 52 | 242 | Crystalstructure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library [unidentified],4Q3K_B Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library [unidentified] |
| 6A6O_A | 5.48e-17 | 256 | 481 | 45 | 282 | ChainA, Esterase/lipase-like protein [Caldicellulosiruptor lactoaceticus 6A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96402 | 2.50e-14 | 257 | 451 | 146 | 367 | Esterase LipC OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipC PE=1 SV=1 |
| Q50681 | 9.12e-14 | 244 | 436 | 159 | 374 | Probable carboxylic ester hydrolase LipM OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipM PE=1 SV=1 |
| I6Y9F7 | 8.56e-12 | 251 | 438 | 154 | 363 | Esterase LipQ OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lipQ PE=1 SV=1 |
| D5EV35 | 1.75e-10 | 267 | 463 | 49 | 258 | Acetylxylan esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axeA1 PE=1 SV=1 |
| Q84LM4 | 4.28e-10 | 250 | 453 | 516 | 741 | Acylamino-acid-releasing enzyme OS=Arabidopsis thaliana OX=3702 GN=AARE PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
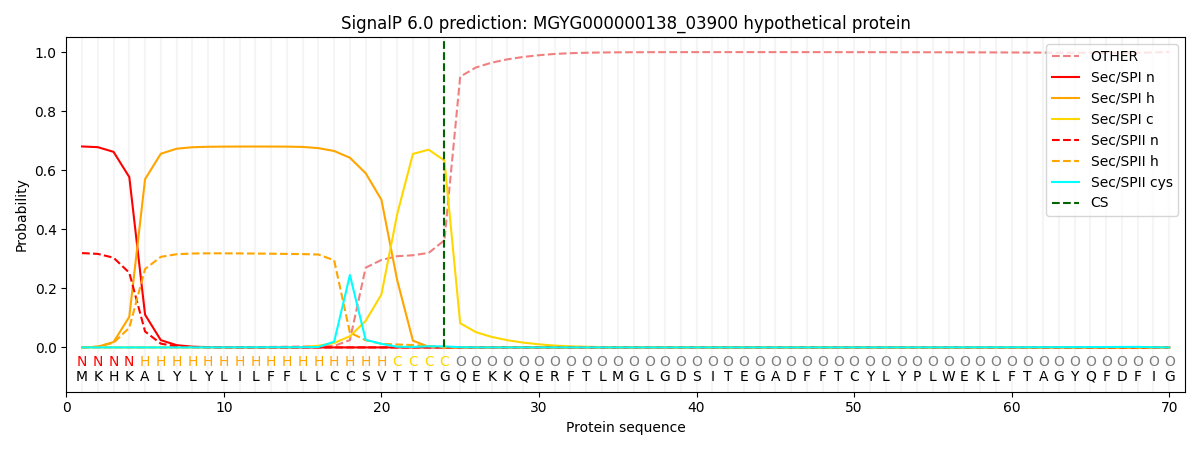
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000835 | 0.670103 | 0.328211 | 0.000314 | 0.000272 | 0.000248 |