You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000139_01484
You are here: Home > Sequence: MGYG000000139_01484
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
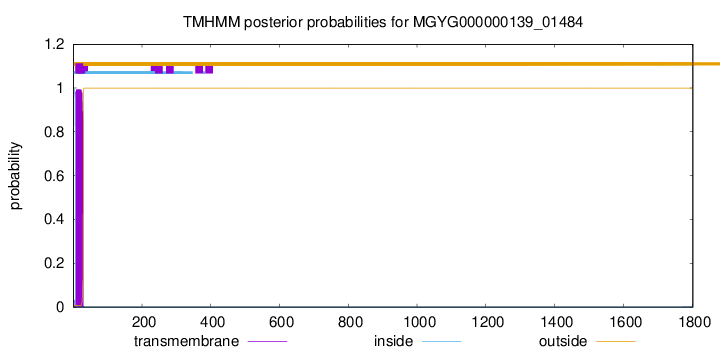
TMHMM annotations
Basic Information help
| Species | Eubacterium_G sp000434315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_G; Eubacterium_G sp000434315 | |||||||||||
| CAZyme ID | MGYG000000139_01484 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 158289; End: 163697 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 40 | 572 | 7.3e-110 | 0.9898167006109979 |
| CBM32 | 881 | 1004 | 1.7e-16 | 0.7903225806451613 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02585 | PIG-L | 3.11e-13 | 1179 | 1251 | 1 | 79 | GlcNAc-PI de-N-acetylase. Members of this family are related to PIG-L an N-acetylglucosaminylphosphatidylinositol de-N-acetylase (EC:3.5.1.89) that catalyzes the second step in GPI biosynthesis. |
| pfam00754 | F5_F8_type_C | 3.01e-09 | 727 | 862 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.60e-08 | 878 | 1015 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| PRK02122 | PRK02122 | 5.01e-07 | 1173 | 1235 | 368 | 430 | glucosamine-6-phosphate deaminase-like protein; Validated |
| COG2120 | LmbE | 2.91e-06 | 1175 | 1249 | 11 | 87 | N-acetylglucosaminyl deacetylase, LmbE family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW76217.1 | 4.49e-249 | 35 | 1039 | 29 | 1048 |
| QIB55915.1 | 4.49e-249 | 35 | 1039 | 29 | 1048 |
| QRO38181.1 | 1.38e-244 | 39 | 1026 | 36 | 1041 |
| QBF74993.1 | 1.38e-244 | 39 | 1026 | 36 | 1041 |
| QRO38183.1 | 7.20e-244 | 578 | 1615 | 24 | 1021 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 3.50e-40 | 39 | 572 | 8 | 574 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 7V6I_A | 3.10e-39 | 31 | 575 | 7 | 609 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 5GQC_A | 1.21e-33 | 37 | 572 | 15 | 594 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 6KQT_A | 1.07e-25 | 32 | 389 | 237 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 4.31e-25 | 32 | 389 | 237 | 617 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
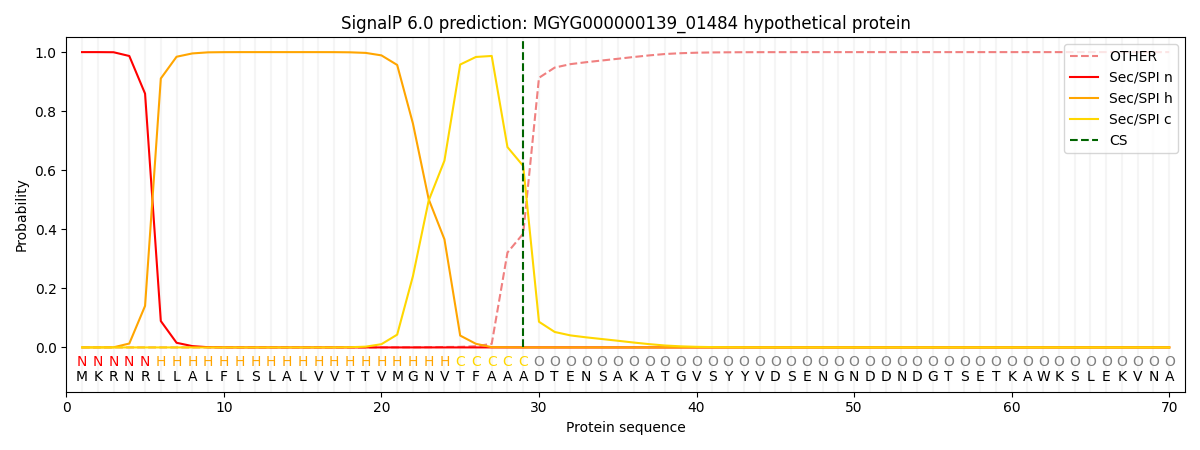
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000271 | 0.999049 | 0.000156 | 0.000190 | 0.000157 | 0.000146 |