You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000139_02375
You are here: Home > Sequence: MGYG000000139_02375
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
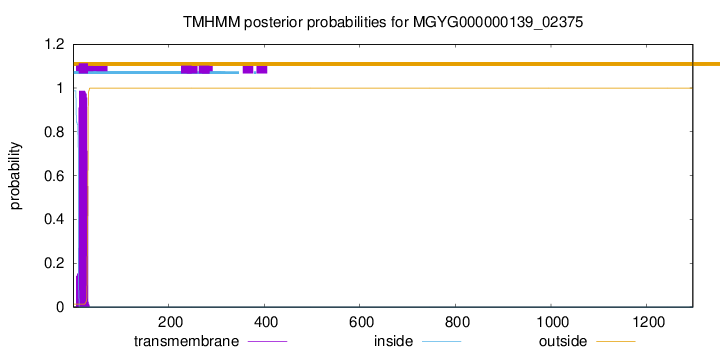
TMHMM annotations
Basic Information help
| Species | Eubacterium_G sp000434315 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_G; Eubacterium_G sp000434315 | |||||||||||
| CAZyme ID | MGYG000000139_02375 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27109; End: 31002 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 99 | 369 | 4.5e-100 | 0.9923954372623575 |
| GH18 | 771 | 1046 | 1.6e-51 | 0.9493243243243243 |
| CBM32 | 628 | 741 | 9.4e-27 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06545 | GH18_3CO4_chitinase | 6.07e-55 | 773 | 1053 | 2 | 253 | The Bacteroides thetaiotaomicron protein represented by pdb structure 3CO4 is an uncharacterized bacterial member of the family 18 glycosyl hydrolases with homologs found in Flavobacterium, Stigmatella, and Pseudomonas. |
| pfam00704 | Glyco_hydro_18 | 6.44e-47 | 773 | 1041 | 3 | 307 | Glycosyl hydrolases family 18. |
| smart00636 | Glyco_18 | 1.11e-43 | 771 | 1041 | 1 | 334 | Glyco_18 domain. |
| cd06548 | GH18_chitinase | 6.03e-42 | 773 | 1041 | 2 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| cd00598 | GH18_chitinase-like | 5.32e-35 | 773 | 1041 | 2 | 210 | The GH18 (glycosyl hydrolase, family 18) type II chitinases hydrolyze chitin, an abundant polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) which is a major component of the cell wall of fungi and the exoskeleton of arthropods. Chitinases have been identified in viruses, bacteria, fungi, protozoan parasites, insects, and plants. The structure of the GH18 domain is an eight-stranded beta/alpha barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. The GH18 family includes chitotriosidase, chitobiase, hevamine, zymocin-alpha, narbonin, SI-CLP (stabilin-1 interacting chitinase-like protein), IDGF (imaginal disc growth factor), CFLE (cortical fragment-lytic enzyme) spore hydrolase, the type III and type V plant chitinases, the endo-beta-N-acetylglucosaminidases, and the chitolectins. The GH85 (glycosyl hydrolase, family 85) ENGases (endo-beta-N-acetylglucosaminidases) are closely related to the GH18 chitinases and are included in this alignment model. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS15177.1 | 7.14e-148 | 1 | 506 | 1 | 498 |
| VEI36204.1 | 3.11e-142 | 9 | 503 | 8 | 483 |
| BCJ97694.1 | 1.76e-117 | 5 | 506 | 2 | 514 |
| QLY78876.1 | 2.17e-108 | 49 | 489 | 43 | 471 |
| QAA34430.1 | 6.27e-106 | 16 | 493 | 8 | 468 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XYZ_A | 3.77e-27 | 782 | 1058 | 29 | 306 | Crystalstructure of the GH18 chitinase ChiB from the chitin utilization locus of Flavobacterium johnsoniae [Flavobacterium johnsoniae UW101] |
| 2RV9_A | 1.58e-24 | 620 | 744 | 8 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 1.63e-24 | 620 | 744 | 9 | 137 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 3.03e-24 | 620 | 744 | 9 | 137 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 2RVA_A | 1.31e-20 | 620 | 744 | 8 | 137 | Solutionstructure of chitosan-binding module 2 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 2.68e-27 | 49 | 305 | 3 | 249 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| Q8NKF9 | 1.07e-18 | 69 | 376 | 37 | 399 | Glucan 1,3-beta-glucosidase OS=Candida oleophila OX=45573 GN=EXG1 PE=3 SV=1 |
| Q9W5U2 | 1.16e-18 | 768 | 980 | 963 | 1210 | Probable chitinase 10 OS=Drosophila melanogaster OX=7227 GN=Cht10 PE=2 SV=2 |
| B0XN12 | 2.35e-18 | 69 | 341 | 42 | 310 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
| B8N151 | 1.20e-17 | 69 | 304 | 31 | 260 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
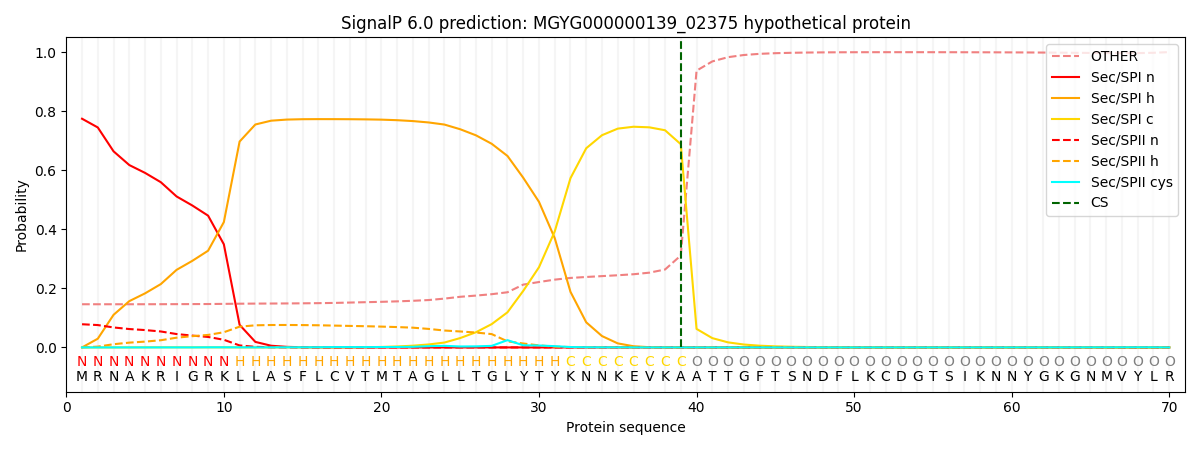
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.153302 | 0.762743 | 0.080475 | 0.002462 | 0.000568 | 0.000426 |