You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000140_00941
You are here: Home > Sequence: MGYG000000140_00941
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
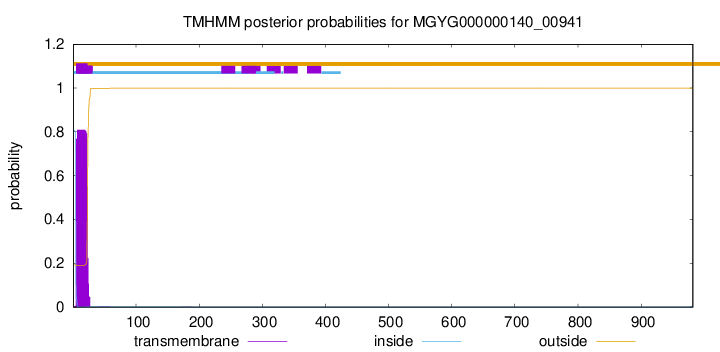
TMHMM annotations
Basic Information help
| Species | UMGS1375 sp900066615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UMGS1375; UMGS1375 sp900066615 | |||||||||||
| CAZyme ID | MGYG000000140_00941 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 383297; End: 386245 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 342 | 674 | 1.3e-16 | 0.8830409356725146 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 3.55e-23 | 83 | 224 | 3 | 150 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| cd15482 | Sialidase_non-viral | 3.62e-13 | 366 | 575 | 42 | 240 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam02368 | Big_2 | 3.97e-11 | 728 | 803 | 3 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| COG5492 | YjdB | 5.24e-10 | 715 | 851 | 171 | 306 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| sd00036 | LRR_3 | 1.95e-09 | 874 | 948 | 63 | 127 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT17714.1 | 3.00e-257 | 42 | 717 | 934 | 1599 |
| ADO53743.1 | 3.58e-257 | 37 | 717 | 939 | 1606 |
| BBA55846.1 | 4.45e-256 | 37 | 717 | 918 | 1585 |
| AXM91086.1 | 7.52e-256 | 37 | 717 | 918 | 1585 |
| QTQ18982.1 | 1.02e-255 | 37 | 717 | 939 | 1606 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P85991 | 5.35e-10 | 646 | 805 | 93 | 257 | Ig-like virion protein OS=Serratia phage KSP90 OX=552528 PE=1 SV=2 |
| P43478 | 5.74e-06 | 721 | 797 | 313 | 388 | Kappa-carrageenase OS=Pseudoalteromonas carrageenovora OX=227 GN=cgkA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
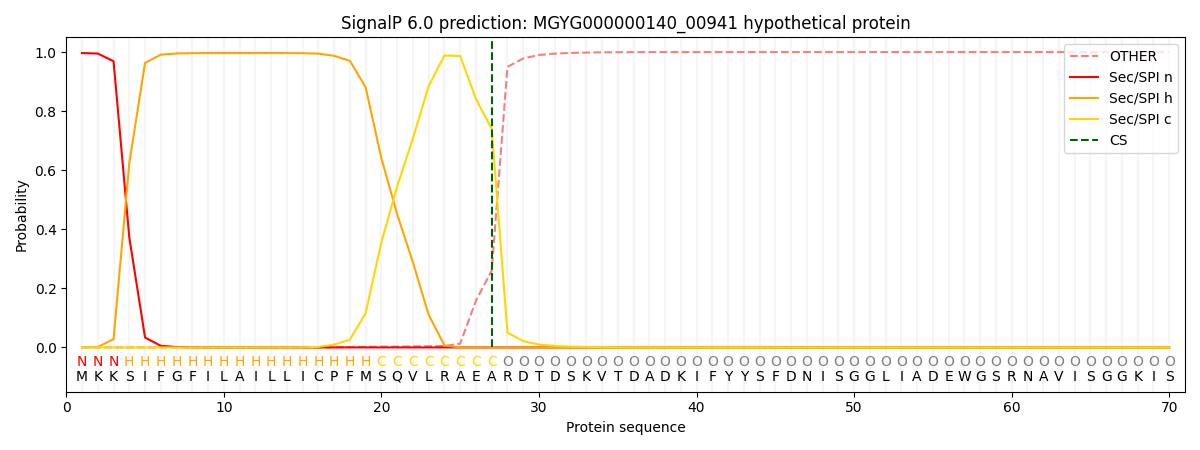
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000509 | 0.994542 | 0.004308 | 0.000236 | 0.000205 | 0.000181 |