You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000140_02984
You are here: Home > Sequence: MGYG000000140_02984
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1375 sp900066615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; UMGS1375; UMGS1375 sp900066615 | |||||||||||
| CAZyme ID | MGYG000000140_02984 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25659; End: 30347 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 845 | 1049 | 7.4e-68 | 0.9675925925925926 |
| CBM6 | 1106 | 1224 | 1.6e-28 | 0.8695652173913043 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1472 | BglX | 5.54e-52 | 846 | 1088 | 59 | 319 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| pfam01915 | Glyco_hydro_3_C | 4.91e-47 | 50 | 277 | 1 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PRK15098 | PRK15098 | 5.38e-40 | 38 | 405 | 383 | 751 | beta-glucosidase BglX. |
| pfam00933 | Glyco_hydro_3 | 4.87e-36 | 846 | 1077 | 65 | 313 | Glycosyl hydrolase family 3 N terminal domain. |
| cd04080 | CBM6_cellulase-like | 6.53e-34 | 1120 | 1224 | 39 | 144 | Carbohydrate Binding Module 6 (CBM6); appended to glycoside hydrolase (GH) domains, including GH5 (cellulase). This family includes carbohydrate binding module 6 (CBM6) domains that are appended to several glycoside hydrolase (GH) domains, including GH5 (cellulase) and GH16, as well as to coagulation factor 5/8 carbohydrate-binding domains. CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. The CBM6s are appended to GHs that display a diversity of substrate specificities. For some members of this family information is available about the specific substrates of the appended GH domains. It includes the CBM domains of various enzymes involved in cell wall degradation including, an extracellular beta-1,3-glucanase from Lysobacter enzymogenes encoded by the gluC gene (its catalytic domain belongs to the GH16 family), the tandem CBM domains of Pseudomonas sp. PE2 beta-1,3(4)-glucanase A (its catalytic domain also belongs to GH16), and a family 6 CBM from Cellvibrio mixtus Endoglucanase 5A (CmCBM6) which binds to the beta1,4-beta1,3-mixed linked glucans lichenan, and barley beta-glucan, cello-oligosaccharides, insoluble forms of cellulose, the beta1,3-glucan laminarin, and xylooligosaccharides, and the CBM6 of Fibrobacter succinogenes S85 XynD xylanase, appended to a GH10 domain, and Cellvibrio japonicas Cel5G appended to a GH5 (cellulase) domain. GH5 (cellulase) family includes enzymes with several known activities such as endoglucanase, beta-mannanase, and xylanase, which are involved in the degradation of cellulose and xylans. GH16 family includes enzymes with lichenase, xyloglucan endotransglycosylase (XET), and beta-agarase activities. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. For CmCBM6 it has been shown that these two binding sites have different ligand specificities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOZ95010.1 | 1.48e-204 | 21 | 1085 | 12 | 803 |
| QIB55852.1 | 2.45e-203 | 36 | 1085 | 26 | 785 |
| QMW76281.1 | 2.45e-203 | 36 | 1085 | 26 | 785 |
| QBE95841.1 | 2.53e-203 | 36 | 1085 | 48 | 807 |
| QAA32414.1 | 2.35e-199 | 38 | 1085 | 28 | 807 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WUG_A | 1.14e-126 | 40 | 1224 | 38 | 955 | Expression,characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii],5WVP_A Expression, characterization and crystal structure of a novel beta-glucosidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 2X42_A | 3.86e-72 | 36 | 410 | 322 | 704 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with alpha-D-glucose [Thermotoga neapolitana DSM 4359] |
| 2X40_A | 3.86e-72 | 36 | 410 | 322 | 704 | Structureof beta-glucosidase 3B from Thermotoga neapolitana in complex with glycerol [Thermotoga neapolitana DSM 4359],2X41_A Structure of beta-glucosidase 3B from Thermotoga neapolitana in complex with glucose [Thermotoga neapolitana DSM 4359] |
| 5WAB_A | 6.47e-53 | 846 | 1108 | 36 | 312 | CrystalStructure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_B Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_C Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703],5WAB_D Crystal Structure of Bifidobacterium adolescentis GH3 beta-glucosidase [Bifidobacterium adolescentis ATCC 15703] |
| 7MS2_A | 5.22e-52 | 844 | 1081 | 34 | 286 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16084 | 6.29e-176 | 36 | 1071 | 26 | 811 | Beta-glucosidase A OS=Butyrivibrio fibrisolvens OX=831 GN=bglA PE=3 SV=1 |
| P15885 | 6.95e-110 | 37 | 1223 | 8 | 902 | Beta-glucosidase OS=Ruminococcus albus OX=1264 PE=3 SV=1 |
| Q5BFG8 | 1.75e-56 | 805 | 1088 | 7 | 293 | Beta-glucosidase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglB PE=1 SV=1 |
| E7CY69 | 2.77e-53 | 846 | 1108 | 36 | 312 | Exo-alpha-(1->6)-L-arabinopyranosidase OS=Bifidobacterium longum OX=216816 GN=apy PE=1 SV=1 |
| F6C6C1 | 2.13e-52 | 846 | 1108 | 36 | 312 | Exo-alpha-(1->6)-L-arabinopyranosidase OS=Bifidobacterium breve (strain ACS-071-V-Sch8b) OX=866777 GN=HMPREF9228_1477 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
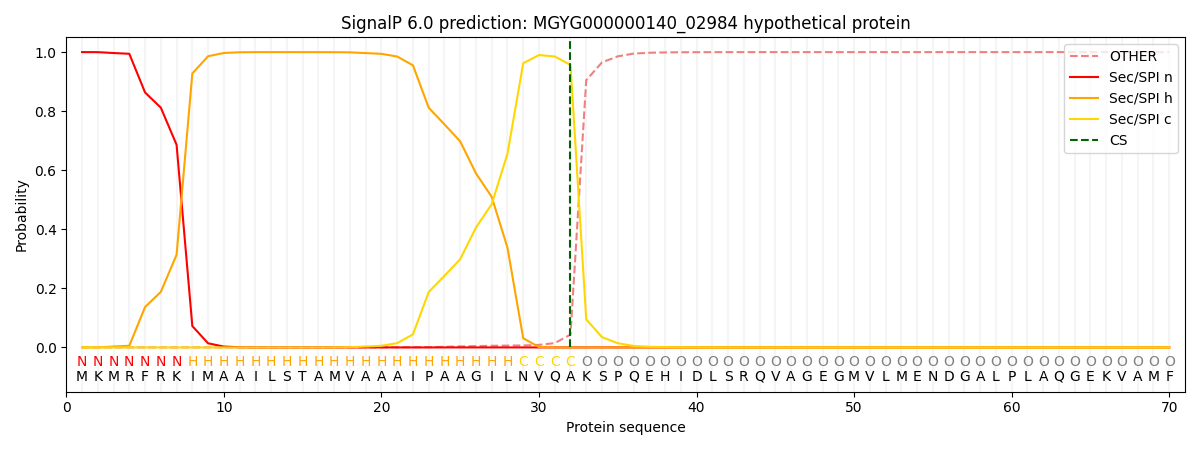
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000471 | 0.998566 | 0.000219 | 0.000321 | 0.000218 | 0.000166 |
