You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000144_00896
You are here: Home > Sequence: MGYG000000144_00896
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Barnesiella intestinihominis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Barnesiellaceae; Barnesiella; Barnesiella intestinihominis | |||||||||||
| CAZyme ID | MGYG000000144_00896 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67242; End: 68267 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 30 | 313 | 1.9e-96 | 0.9822695035460993 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.20e-42 | 38 | 314 | 11 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.43e-21 | 30 | 337 | 49 | 391 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT79278.1 | 1.18e-179 | 15 | 341 | 20 | 345 |
| QDM10317.1 | 1.18e-179 | 15 | 341 | 20 | 345 |
| CBK67193.1 | 9.66e-179 | 15 | 341 | 20 | 345 |
| QGT72468.1 | 1.37e-178 | 15 | 341 | 20 | 345 |
| QDH53451.1 | 7.91e-178 | 15 | 341 | 20 | 345 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EE9_A | 2.01e-79 | 30 | 336 | 6 | 306 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
| 7P6G_A | 5.70e-79 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],7P6G_B Chain B, Endoglucanase [uncultured bacterium],7P6H_A Chain A, Endoglucanase [uncultured bacterium],7P6H_B Chain B, Endoglucanase [uncultured bacterium] |
| 6ZZ3_A | 7.81e-79 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],6ZZ3_B Chain B, Endoglucanase [uncultured bacterium],6ZZ3_C Chain C, Endoglucanase [uncultured bacterium],6ZZ3_D Chain D, Endoglucanase [uncultured bacterium] |
| 7P6I_A | 8.05e-79 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],7P6J_A Chain A, Endoglucanase [uncultured bacterium],7P6J_B Chain B, Endoglucanase [uncultured bacterium],7P6J_C Chain C, Endoglucanase [uncultured bacterium],7P6J_D Chain D, Endoglucanase [uncultured bacterium] |
| 5LJF_A | 1.61e-78 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],5LJF_B Chain B, Endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P58599 | 5.65e-56 | 30 | 337 | 119 | 423 | Endoglucanase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=egl PE=3 SV=1 |
| P17974 | 4.53e-55 | 30 | 337 | 121 | 425 | Endoglucanase OS=Ralstonia solanacearum OX=305 GN=egl PE=1 SV=2 |
| Q96WQ8 | 1.48e-44 | 29 | 324 | 35 | 318 | Probable endo-beta-1,4-glucanase B OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=eglB PE=3 SV=1 |
| Q2UPQ4 | 2.97e-44 | 30 | 324 | 35 | 318 | Probable endo-beta-1,4-glucanase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=eglB PE=3 SV=1 |
| B8MW97 | 2.97e-44 | 30 | 324 | 35 | 318 | probable endo-beta-1,4-glucanase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
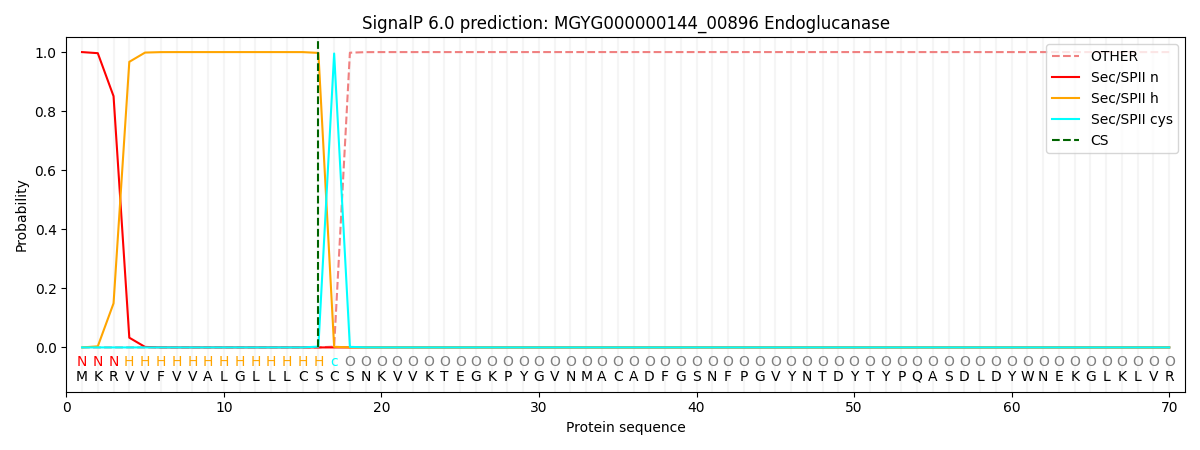
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000056 | 0.000000 | 0.000000 | 0.000000 |
