You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000147_01021
You are here: Home > Sequence: MGYG000000147_01021
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
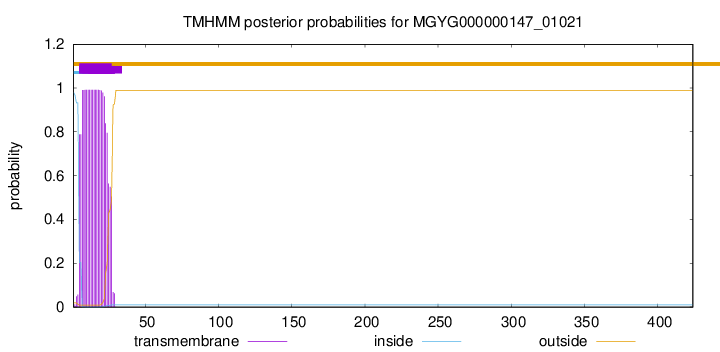
TMHMM annotations
Basic Information help
| Species | Bacillus paralicheniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus paralicheniformis | |||||||||||
| CAZyme ID | MGYG000000147_01021 | |||||||||||
| CAZy Family | GH53 | |||||||||||
| CAZyme Description | Arabinogalactan endo-beta-1,4-galactanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 100985; End: 102259 Strand: + | |||||||||||
Full Sequence Download help
| MKNVVAVFVV LIVVLGALGT GGPAEAARGS KAAESGLFVE KVSGLHKDFI KGVDVSSIIA | 60 |
| LEESGVAFYN ESGKKQDIFK TLKEAGVNYV RVRIWNDPYD ANGNGYGGGN NDLKKAIQIG | 120 |
| KRATANGMKL LADFHYSDFW ADPAKQKAPK AWANLNFEDK KTALYQYTKQ SLEAMKAAGI | 180 |
| NVGMVQVGNE TNGGLAGETD WAKMSQLFNA GSQAVRETDS NILVALHFTN PETSGRYAWI | 240 |
| AETLHRHHVD YDVFASSYYP FWHGTLKNLT SVLTSVADTY GKKVMVAETS YTYTAEDGDG | 300 |
| HENTAPKNGQ TLNYPVTVQG QANAVRDVIQ AVSDVGEAGI GVFYWEPAWI PVGPARQLEK | 360 |
| NKALWETYGS GWATSYAAEY DPEDAGKWFG GSAVDNQALF DFKGRPLPSL NVFKYVDTGT | 420 |
| PFKN | 424 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 50 | 413 | 6.8e-138 | 0.9970760233918129 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3867 | GanB | 0.0 | 32 | 423 | 22 | 401 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
| pfam07745 | Glyco_hydro_53 | 2.04e-167 | 50 | 413 | 1 | 333 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| COG2730 | BglC | 7.81e-05 | 75 | 345 | 75 | 357 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AJO20700.1 | 3.98e-315 | 1 | 424 | 1 | 424 |
| QFY40674.1 | 3.98e-315 | 1 | 424 | 1 | 424 |
| QEO05120.1 | 3.98e-315 | 1 | 424 | 1 | 424 |
| ARA87839.1 | 3.98e-315 | 1 | 424 | 1 | 424 |
| AYQ18610.1 | 3.98e-315 | 1 | 424 | 1 | 424 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R8L_A | 4.21e-287 | 28 | 424 | 3 | 399 | Thestructure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1R8L_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1UR0_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR0_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],2CCR_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2CCR_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis] |
| 2GFT_A | 3.45e-286 | 28 | 424 | 3 | 399 | ChainA, Glycosyl Hydrolase Family 53 [Bacillus licheniformis],2GFT_B Chain B, Glycosyl Hydrolase Family 53 [Bacillus licheniformis] |
| 7OSK_A | 2.44e-87 | 37 | 421 | 38 | 399 | ChainA, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230],7OSK_B Chain B, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230] |
| 1HJS_A | 7.69e-44 | 51 | 350 | 5 | 301 | Structureof two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_A Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus] |
| 1HJQ_A | 5.69e-43 | 51 | 350 | 5 | 301 | Structureof two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q65CX5 | 9.78e-301 | 1 | 424 | 1 | 424 | Endo-beta-1,4-galactanase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=ganB PE=1 SV=1 |
| O07013 | 1.29e-235 | 1 | 419 | 1 | 423 | Endo-beta-1,4-galactanase OS=Bacillus subtilis (strain 168) OX=224308 GN=ganB PE=1 SV=1 |
| P48843 | 9.42e-65 | 49 | 414 | 6 | 347 | Uncharacterized protein in bgaB 5'region (Fragment) OS=Niallia circulans OX=1397 PE=3 SV=1 |
| P48841 | 6.42e-50 | 49 | 366 | 27 | 345 | Arabinogalactan endo-beta-1,4-galactanase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=ganB PE=1 SV=1 |
| P83692 | 4.21e-43 | 51 | 350 | 5 | 301 | Arabinogalactan endo-beta-1,4-galactanase OS=Thermothelomyces thermophilus OX=78579 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
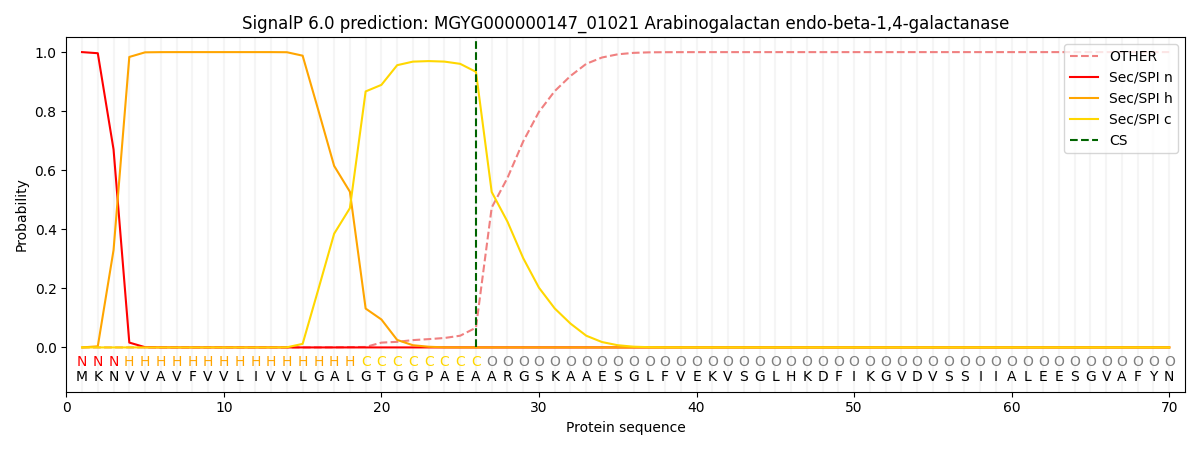
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000187 | 0.999237 | 0.000142 | 0.000142 | 0.000125 | 0.000123 |