You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000147_03810
You are here: Home > Sequence: MGYG000000147_03810
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
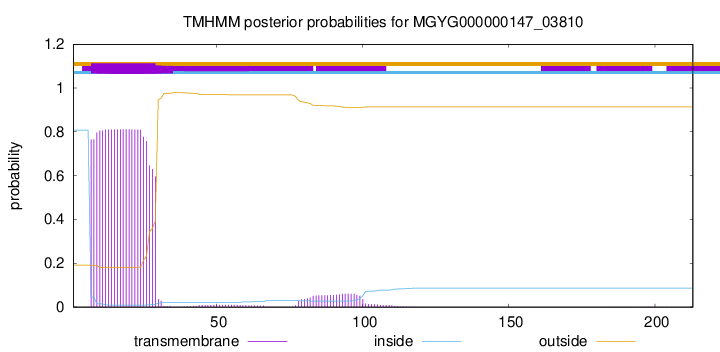
TMHMM annotations
Basic Information help
| Species | Bacillus paralicheniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus paralicheniformis | |||||||||||
| CAZyme ID | MGYG000000147_03810 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52985; End: 53626 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 32 | 211 | 6.7e-71 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00457 | Glyco_hydro_11 | 2.15e-87 | 30 | 209 | 1 | 175 | Glycosyl hydrolases family 11. |
| PRK06215 | PRK06215 | 0.003 | 1 | 136 | 8 | 167 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACF05486.1 | 3.33e-160 | 1 | 213 | 1 | 213 |
| QSF97430.1 | 3.33e-160 | 1 | 213 | 1 | 213 |
| VEB17655.1 | 3.33e-160 | 1 | 213 | 1 | 213 |
| ARA84517.1 | 3.33e-160 | 1 | 213 | 1 | 213 |
| ATJ03481.1 | 3.33e-160 | 1 | 213 | 1 | 213 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XXN_A | 1.90e-129 | 29 | 213 | 1 | 185 | Crystalstructure of a mesophilic xylanase A from Bacillus subtilis 1A1 [Bacillus subtilis],2DCY_A Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_B Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_C Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_D Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_E Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis] |
| 1BVV_A | 5.45e-129 | 29 | 213 | 1 | 185 | ChainA, ENDO-1,4-BETA-XYLANASE [Niallia circulans],1XNB_A Chain A, XYLANASE [Niallia circulans],2B42_B Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase [Bacillus subtilis],3HD8_B Crystal structure of the Triticum aestivum xylanase inhibitor-IIA in complex with bacillus subtilis xylanase [Bacillus subtilis],3HD8_D Crystal structure of the Triticum aestivum xylanase inhibitor-IIA in complex with bacillus subtilis xylanase [Bacillus subtilis] |
| 2QZ3_A | 1.56e-128 | 29 | 213 | 1 | 185 | ChainA, Endo-1,4-beta-xylanase A [Bacillus subtilis],2QZ3_B Chain B, Endo-1,4-beta-xylanase A [Bacillus subtilis],2Z79_A Chain A, Endo-1,4-beta-xylanase A [Bacillus subtilis],2Z79_B Chain B, Endo-1,4-beta-xylanase A [Bacillus subtilis] |
| 1HV0_A | 2.22e-128 | 29 | 213 | 1 | 185 | ChainA, ENDO-1,4-BETA-XYLANASE [Niallia circulans] |
| 2BVV_A | 2.22e-128 | 29 | 213 | 1 | 185 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE) [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P18429 | 7.11e-147 | 1 | 213 | 1 | 213 | Endo-1,4-beta-xylanase A OS=Bacillus subtilis (strain 168) OX=224308 GN=xynA PE=1 SV=1 |
| P09850 | 2.04e-146 | 1 | 213 | 1 | 213 | Endo-1,4-beta-xylanase OS=Niallia circulans OX=1397 GN=xlnA PE=1 SV=1 |
| V9TXH2 | 5.19e-121 | 1 | 213 | 1 | 210 | Endo-1,4-beta-xylanase Xyn11E OS=Paenibacillus barcinonensis OX=198119 GN=xyn11E PE=1 SV=1 |
| P45705 | 6.05e-120 | 3 | 213 | 2 | 210 | Endo-1,4-beta-xylanase A OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=3 SV=2 |
| Q0CFS3 | 6.79e-77 | 33 | 213 | 56 | 231 | Probable endo-1,4-beta-xylanase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xlnA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
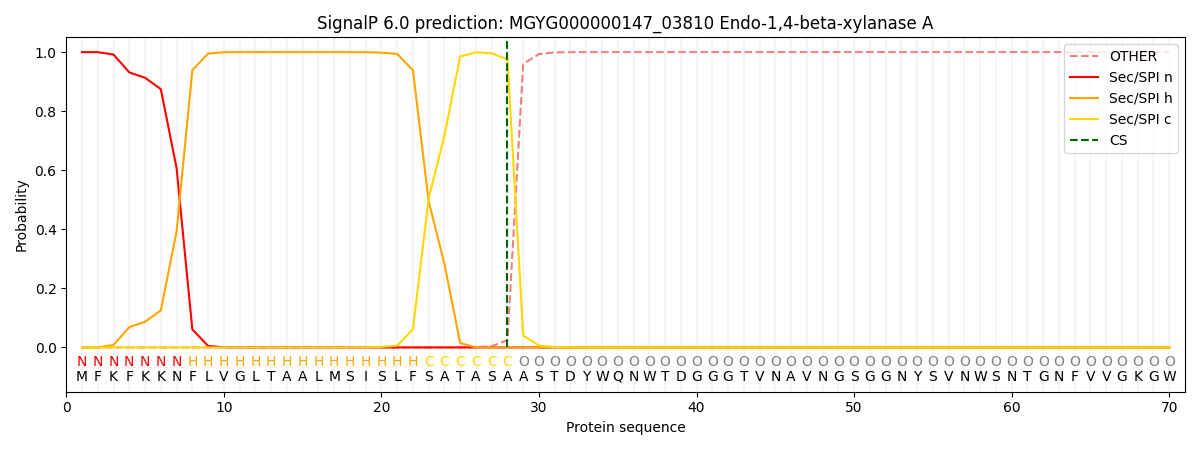
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000224 | 0.999054 | 0.000168 | 0.000205 | 0.000160 | 0.000146 |