You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000147_04314
You are here: Home > Sequence: MGYG000000147_04314
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
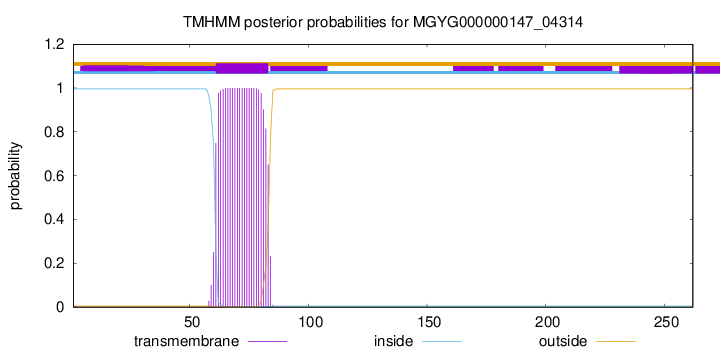
TMHMM annotations
Basic Information help
| Species | Bacillus paralicheniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; Bacillus paralicheniformis | |||||||||||
| CAZyme ID | MGYG000000147_04314 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12407; End: 13195 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033598 | elast_bind_EbpS | 6.00e-10 | 110 | 260 | 296 | 466 | elastin-binding protein EbpS. The elastin-binding protein EbpS is an adhesin described in Staphylococcus aureus, with orthologs found in many additional staphylococcal species. EbpS is a membrane protein that lacks an N-terminal signal peptide region, has extensive regions low-complexity sequence rich in Asn and Gln, and has a C-terminal LysM domain. |
| pfam01476 | LysM | 2.39e-07 | 215 | 260 | 1 | 43 | LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known. |
| PRK00409 | PRK00409 | 5.33e-05 | 115 | 260 | 525 | 658 | recombination and DNA strand exchange inhibitor protein; Reviewed |
| PTZ00144 | PTZ00144 | 1.17e-04 | 161 | 237 | 127 | 203 | dihydrolipoamide succinyltransferase; Provisional |
| PRK13914 | PRK13914 | 1.51e-04 | 168 | 259 | 149 | 243 | invasion associated endopeptidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEB19547.1 | 2.96e-116 | 1 | 262 | 1 | 262 |
| BCE07233.1 | 4.13e-109 | 1 | 262 | 1 | 262 |
| BCE09033.1 | 4.13e-109 | 1 | 262 | 1 | 262 |
| ARA86213.1 | 1.24e-103 | 21 | 262 | 1 | 242 |
| QFY38387.1 | 1.24e-103 | 21 | 262 | 1 | 242 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P50731 | 1.30e-27 | 1 | 261 | 1 | 237 | Uncharacterized protein YpbE OS=Bacillus subtilis (strain 168) OX=224308 GN=ypbE PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999705 | 0.000265 | 0.000022 | 0.000001 | 0.000001 | 0.000014 |