You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000148_01553
You are here: Home > Sequence: MGYG000000148_01553
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
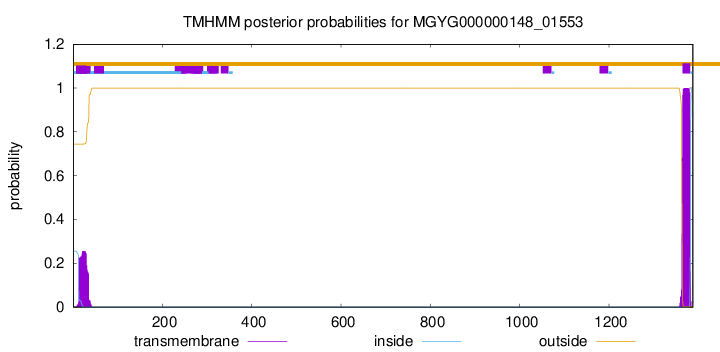
TMHMM annotations
Basic Information help
| Species | Faecalimonas phoceensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas phoceensis | |||||||||||
| CAZyme ID | MGYG000000148_01553 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66066; End: 70232 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 451 | 1114 | 1.9e-213 | 0.8407202216066482 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14498 | Glyco_hyd_65N_2 | 2.11e-29 | 452 | 598 | 100 | 233 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
| pfam14498 | Glyco_hyd_65N_2 | 6.71e-17 | 68 | 185 | 3 | 103 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
| TIGR02169 | SMC_prok_A | 7.63e-06 | 1190 | 1324 | 342 | 497 | chromosome segregation protein SMC, primarily archaeal type. SMC (structural maintenance of chromosomes) proteins bind DNA and act in organizing and segregating chromosomes for partition. SMC proteins are found in bacteria, archaea, and eukaryotes. It is found in a single copy and is homodimeric in prokaryotes, but six paralogs (excluded from this family) are found in eukarotes, where SMC proteins are heterodimeric. This family represents the SMC protein of archaea and a few bacteria (Aquifex, Synechocystis, etc); the SMC of other bacteria is described by TIGR02168. The N- and C-terminal domains of this protein are well conserved, but the central hinge region is skewed in composition and highly divergent. [Cellular processes, Cell division, DNA metabolism, Chromosome-associated proteins] |
| PRK01156 | PRK01156 | 2.23e-05 | 1202 | 1327 | 465 | 597 | chromosome segregation protein; Provisional |
| TIGR04523 | Mplasa_alph_rch | 4.83e-04 | 1166 | 1328 | 537 | 694 | helix-rich Mycoplasma protein. Members of this family occur strictly within a subset of Mycoplasma species. Members average 750 amino acids in length, including signal peptide. Sequences are predicted (Jpred 3) to be almost entirely alpha-helical. These sequences show strong periodicity (consistent with long alpha helical structures) and low complexity rich in D,E,N,Q, and K. Genes encoding these proteins are often found in tandem. The function is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM13266.1 | 1.42e-279 | 41 | 1154 | 40 | 850 |
| ASM69030.1 | 4.37e-255 | 11 | 1154 | 6 | 847 |
| QHB24557.1 | 1.04e-218 | 458 | 1152 | 175 | 851 |
| QEI32060.1 | 1.04e-218 | 458 | 1152 | 175 | 851 |
| QRT30709.1 | 3.24e-218 | 458 | 1152 | 175 | 851 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2EAB_A | 6.49e-207 | 409 | 1155 | 116 | 895 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
| 2EAD_A | 5.00e-206 | 409 | 1155 | 116 | 895 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum],2EAD_B Chain B, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2EAE_A | 9.57e-206 | 409 | 1155 | 115 | 894 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2RDY_A | 4.18e-91 | 458 | 1157 | 111 | 792 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 4UFC_A | 4.94e-91 | 452 | 1114 | 125 | 743 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 2.46e-73 | 458 | 1099 | 162 | 799 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| A2R797 | 3.52e-61 | 449 | 1100 | 127 | 764 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q5AU81 | 1.33e-52 | 457 | 1113 | 140 | 797 | Alpha-fucosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=afcA PE=1 SV=1 |
| Q2USL3 | 2.36e-37 | 457 | 1099 | 129 | 701 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
| P0DTR5 | 2.24e-08 | 192 | 426 | 836 | 1060 | A type blood alpha-D-galactosamine galactosaminidase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
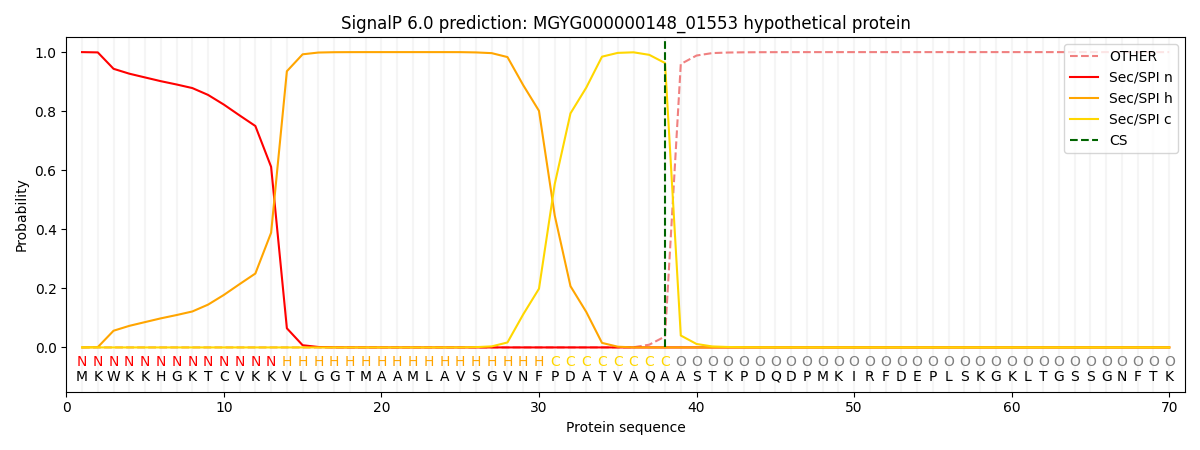
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000352 | 0.998867 | 0.000203 | 0.000223 | 0.000182 | 0.000149 |