You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000148_01573
You are here: Home > Sequence: MGYG000000148_01573
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
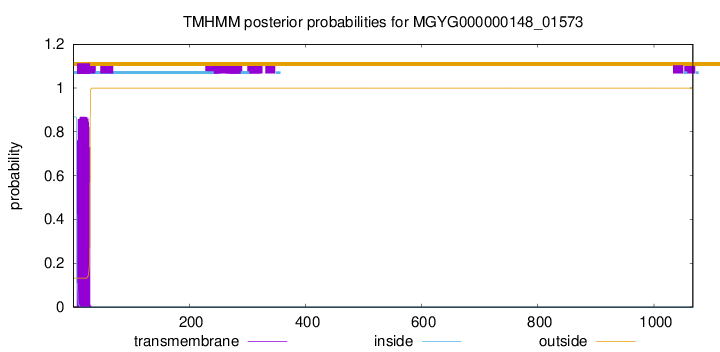
TMHMM annotations
Basic Information help
| Species | Faecalimonas phoceensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Faecalimonas; Faecalimonas phoceensis | |||||||||||
| CAZyme ID | MGYG000000148_01573 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9552; End: 12755 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 43 | 547 | 3.2e-128 | 0.9978070175438597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 6.58e-21 | 43 | 391 | 4 | 354 | O-Glycosyl hydrolase family 30. |
| COG5520 | XynC | 5.63e-16 | 183 | 548 | 124 | 427 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam17996 | CE2_N | 5.69e-04 | 607 | 676 | 18 | 83 | Carbohydrate esterase 2 N-terminal. This is the N-terminal beta-sheet domain with jelly roll topology found in CE2 acetyl-esterase from the bacterium Clostridium thermocellum. This enzyme displays dual activities, it catalyses the deacetylation of plant polysaccharides and also potentiates the activity of its appended cellulase catalytic module through its noncatalytic cellulose binding function. This N-terminal jelly-roll domain appears to extend the substrate/cellulose binding cleft of the catalytic domain in C.thermocellum. |
| pfam07532 | Big_4 | 0.001 | 708 | 759 | 5 | 59 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWY99102.1 | 0.0 | 1 | 1028 | 1 | 1044 |
| QIB57138.1 | 0.0 | 1 | 1006 | 1 | 1012 |
| QMW80085.1 | 0.0 | 1 | 1006 | 1 | 1012 |
| ADL05173.1 | 7.74e-293 | 41 | 1054 | 42 | 1308 |
| QJU15030.1 | 1.05e-150 | 116 | 1054 | 302 | 1094 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 2.47e-12 | 45 | 554 | 10 | 501 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 4QAW_A | 2.33e-10 | 112 | 556 | 11 | 395 | Structureof modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_B Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_C Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_D Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_E Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_F Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_G Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_H Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 4FMV_A | 3.39e-10 | 160 | 548 | 66 | 385 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A401ETL2 | 6.57e-31 | 9 | 857 | 11 | 955 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
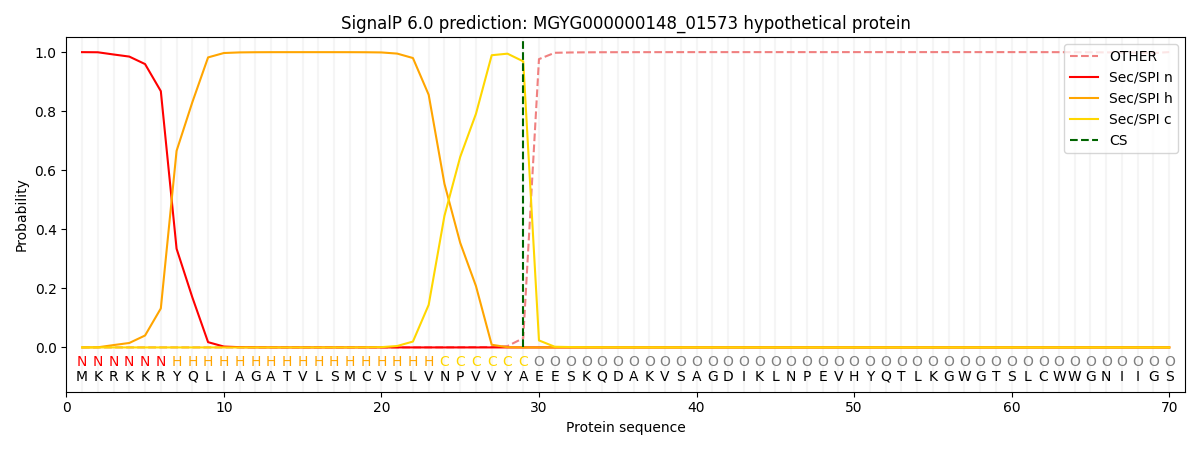
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.003694 | 0.994166 | 0.000611 | 0.000583 | 0.000478 | 0.000433 |